Research relating to machine learning algorithms, including convolutional neural networks, has increased during the past 5 years. The aim of this pilot study was to investigate how accurately a convolutional neural network, trained on Swedish registry data, could perform in predicting cutaneous invasive and in situ melanoma (CMM) within 5 years. A cohort of 1,208,393 individuals was used. Registry data ranged from 4 July 2005 to 31 December 2011, predicting CMM between 1 January 2012 and 31 December 2016. A convolutional neural network with one-dimensional convolutions with respect to time was trained using healthcare databases and registers. The algorithm was trained on 23,886 individuals. Validation was performed on a holdout validation set including 6,000 individuals. After training and validation, the convolutional neural network was evaluated on a test set (1,000 individuals with an CMM occurring within 5 years and 5,000 without). The area under the receiver-operating characteristic curve was 0.59 (95% confidence interval (95% CI) 0.57–0.61). The point on the receiver-operating characteristic curve where sensitivity equalled specificity had a value of 56% (sensitivity 95% CI 53–60% and specificity 95% CI 55–58%). Albeit at an early stage, this pilot investigation demonstrates potential usefulness for machine learning algorithms in predicting melanoma risk.
Key words: area under curve; deep learning; epidemiological methods; machine learning; melanoma; receiver-operating characteristic curve; sensitivity and specificity.
Accepted Jun 27, 2022; Epub ahead of print Jun 27, 2022
Acta Derm Venereol 2022; 102: adv00750.
DOI: 10.2340/actadv.v102.2028
Corr: Sam Polesie, Department of Dermatology and Venereology, Institute of Clinical Sciences, Sahlgrenska Academy, University of Gothenburg, SE-413 45 Gothenburg, Sweden. E-mail: sam.polesie@vgregion.se
SIGNIFICANCE
For all Swedish citizens extensive healthcare data are available in several registries and databases. In this proof of concept study, this data was used to train a machine learning model to predict future risk of melanoma, a potentially lethal skin tumour. Registry data ranging from 2005 to 2011 was used to predict risk of melanoma in the period 2012 to 2016. Using merely this data-set, the machine learning algorithm achieved significantly better than chance alone. While the model needs to be improved and refined in upcoming investigations, this study demonstrates potential usefulness for machine learning in this setting.
INTRODUCTION
Machine learning (ML) algorithms including convolutional neural networks (CNNs) have recently pervaded every aspect of medical imaging, and noteworthy advancements have been made in several medical fields, including radiology (1), dermatology (2–5) and pathology (6). Furthermore, significant progress has been made using neural networks in domains other than image analysis, such as prediction models for specific diseases using available electronic healthcare data and registries. The concept of using neural networks on electronic healthcare data for prediction models, often referred to as computational phenotyping, was first published in 2016 by Cheng et al. (7). To date, only a few investigations have been performed to predict specific dermatological outcomes (8). In future healthcare, it is expected that prediction algorithms will be important to target the increased demands for precision medicine. Moreover, ML algorithms may prove particularly useful for targeted population screening, where high-risk individuals could be identified and automatically invited to screening visits, based on available data in healthcare registries. Finally, these models may reduce healthcare costs and prove to be time efficient.
For a Swedish population, setting up a risk prediction model for cutaneous malignant melanoma (CMM) would be appealing, since the incidence is one of the highest in the world (9). The incidence of CMM has increased dramatically in the Nordic population during the past 3 decades. In Sweden, approximately 500 individuals die from advanced melanoma disease annually. In the early stages, CMM is treatable with an overall good prognosis, whereas in more advanced stages, the prognosis is poor and treatment is costly (10, 11). From a societal perspective, finding novel and complementary tools to identify individuals at higher risk of skin cancer in general and for CMM in particular is a high priority.
Sweden has a public healthcare system, which is largely documented with nationwide registers. This is a world-unique source of information, which, unlike many other countries, is completely disconnected from insurance and compensation systems. The aim of this pilot study was to investigate how accurately a CNN trained on Swedish registry data could perform in predicting CMM within 5 years in individuals without a previous history of CMM.
MATERIALS AND METHODS
Cohort description
This retrospective investigation included individuals from 8 cohorts used in previous research projects. This previous research primarily investigated patients with psoriasis and their respective controls. When merging all of these, the original source cohort consisted of approximately 1.7 million individuals. One of the 8 original cohorts (i.e. patients with psoriasis) has been used in 2 previous investigations (12, 13). A detailed description of all 8 cohorts is presented in Appendix S1. To exclude any systematic bias (i.e. to avoid an over-representation of patients with psoriasis), we only included the 4 control cohorts in our analyses. When merging all these data, 1,489,519 individuals were available.
The registry data ranged from 4 July 2005 to 31 December 2011, predicting CMM between, 1 January 2012 and 31 December 2016. The data comprised time-independent (age, sex, origin, country of birth, income and educational level) and time-dependent variables, including drug prescription (Anatomical Therapeutic Chemical (ATC) and diagnosis (International Classification of Diseases 10th Revision (ICD-10)) codes (Tables I and II). A brief overview of the registries and databases used is shown in Appendix S2.


Included individuals needed to be ≥ 18 years on 4 July 2005 and were excluded if they were ≥ 95 years during 2012. All individuals had to be alive on 31 December 2011. No migration events between 4 July 2005 and 31 December 2016 were allowed. All individuals with a history of CMM (including melanoma in situ) before 1 January 2012 were excluded. After exclusions, the final cohort comprised 1,208,393 individuals in the age range 25–94 years. All these individuals were drawn from the general population, and constituted approximately 18% of the Swedish population within the same age range (i.e. 6.7 million individuals) (14).
Randomization of training, validation, and test-set
All individuals with an occurring CMM in the time-period 1 January 2012 to 31 December 2016 were identified (n = 5,981). To each such individual, 5 age- and sex-matched individuals were drawn randomly from the available controls (n = 29,905). The controls did not develop CMM in the same time-period. All included individuals (n = 35,886) were randomized into a training, validation, and test-set (Fig. 1). The default random number generator was used (Mersenne-Twister) in R version 3.5.3 (https://www.r-project.org/) for the randomization process.

The study was reviewed and approved by the Swedish Ethical Review Authority (registration number 2020-06761) and the Ethics Review Appeals Board (registration number Ö14-2021/3.1).
Outcome
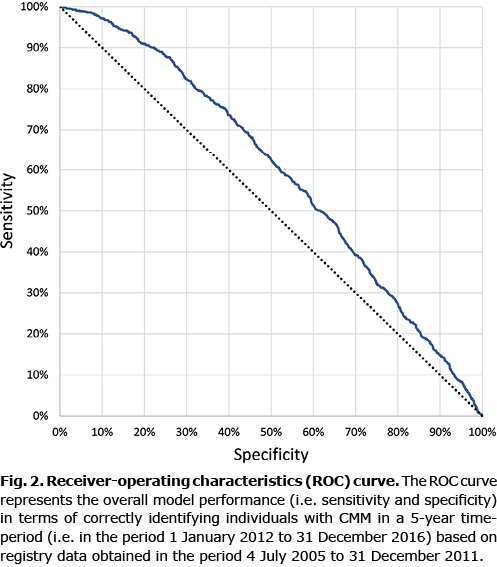
The primary outcome was to investigate at what sensitivity and specificity level a CNN, trained on Swedish registry data, could predict which individuals would be diagnosed with CMM within 5 years. A receiver-operating characteristics curve (ROC) was used to demonstrate the sensitivity and specificity for correctly identifying individuals that would develop CMM. The area under the ROC (AUC) was used to assess the overall model performance.
Model architecture
Different models were trained and validated on the holdout validation set after each epoch. All models were trained using healthcare databases and registers (Appendices S3 and S4). The number of dense layers in models varied from 1 to 5. Within dense layers the number of nodes varied from 64 to 1,024. Models with 1 and 2 convolutional layers (with strides) were trained and the number of filters within the convolutional layers were 4 or 8. In total, 68 different models were trained. Finally, a primary model was selected that achieved maximum performance on the holdout validation set. Only the primary model, along with 9 variations of the model (post-hoc analyses; Table III) were evaluated on the test-set.
The primary model used 1 convolutional layer with 4 filters and a kernel of size (1, 351); i.e. 351 weeks. There were 3 dense layers with 512, 256 and 128 nodes in each, respectively (Fig. S1).
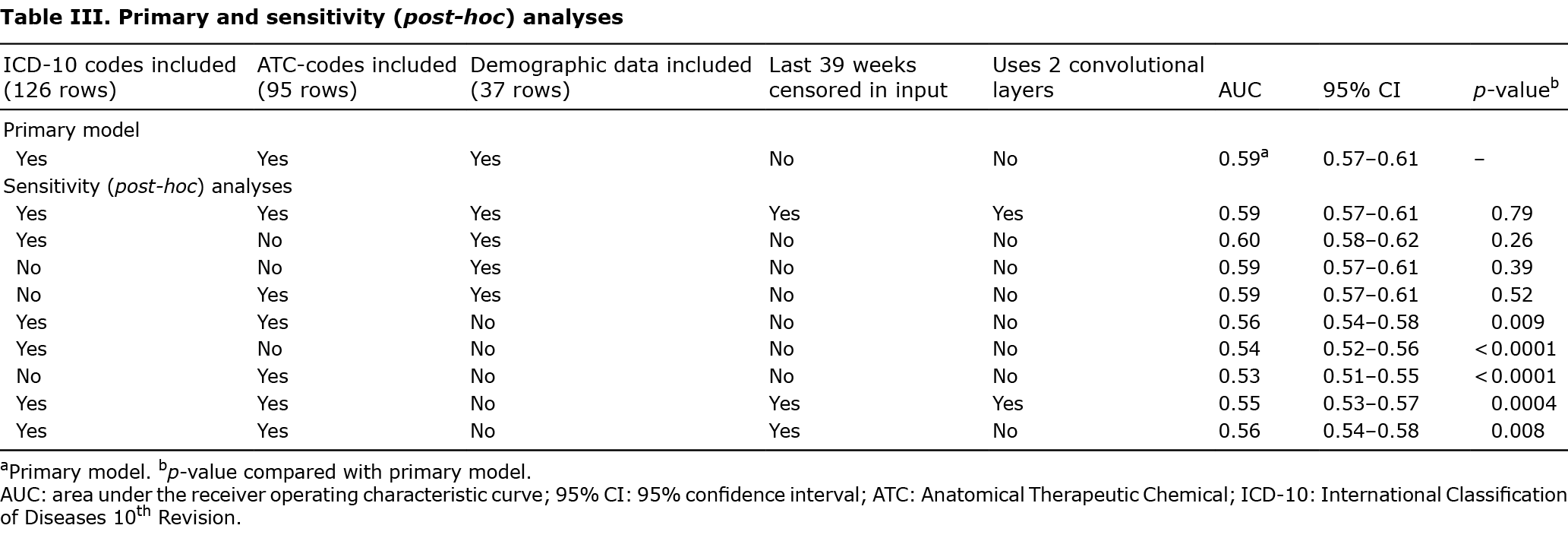
Sensitivity (post-hoc) analyses
To evaluate which data points were important for the model’s AUC, 9 sensitivity (post-hoc) analyses were conducted, which systematically omitted ICD-10 codes and/or ATC-codes, and/or demographic factors, respectively. Models that used 2 convolutional layers were also used, in which the first layer used kernels of size (1, 52) using strides; i.e. 1 year. The second convolutional layer had kernels of size (1, 6); i.e. 6 years. This implied that the last 39 (351-52•6) weeks of the time-dependent variables were censored (i.e. ICD-10 and ATC codes) (Appendices S5 and S6).
Statistical analysis
All data were analysed using R version 3.5.3 (https://www. r-project.org/). All tests were 2-sided and p < 0.05 was considered statistically significant. DeLong’s test for 2 correlated ROC curves was used to compare the performance of different models. Fisher’s exact test and Wilcoxon’s rank sum test were used for 2-sample comparisons.
Hardware and software
The Keras library (version 2.3.1) using the Tensorflow backend (version 1.14.0) was used running on Python version 3.6.9. Model construction was performed using R version 3.5.3 (https://www. r-project.org/) and the R-package Keras was used to call Python and its above libraries. The computer running the training was using the central processing unit (CPU) version on the Keras/Tensorflow routines. The CPU used was an Intel Xeon W-2135 @ 3.7 GHz, with 128 GB random-access memory (RAM). The training of the primary model (147 epochs) took 1 h and 11 min (Fig. S2).
RESULTS
Overall, median age at baseline was 65 years and there was a slight predominance of females (Table IV). The mean number of outpatient ICD-10 diagnoses before 1 January 2012 among all individuals was 14.0 (95% CI 13.7–14.3). The individuals with CMM had a mean of 15.2 diagnoses (95% CI 14.4–16.0) and the corresponding number for the controls was 13.8 (95% CI 13.5–14.1, p < 0.0001). The mean number of inpatient ICD-10 diagnoses before 1 January 2012 among all individuals was 7.4 (95% CI 7.3–7.6). The individuals with CMM had a mean of 6.9 diagnoses (95% CI 6.6–7.2) and the corresponding number for the controls was 7.5 (95% CI 7.4–7.7, p = 0.069). Individuals without CMM had dispensed more pharmaceutical drugs compared with individuals who developed CMM (Table IV).

Of individuals with CMM, 5,786 (96.7%) originated from Nordic countries. The corresponding number among individuals without CMM was 27,212 (91.0%, p < 0.0001) (Table V). Among all 29,905 individuals who did not develop CMM in the time-period, 3,359 (11.2%) had no available ICD-10 diagnoses. The corresponding value among individuals who developed CMM (n = 5,981) was 564 (9.4%, p < 0.0001). Overall, 355 (5.9%) and 2,450 (8.2%) of the individuals with and without CMM died during the observation period (p < 0.0001). For individuals who developed CMM, the median time (interquartile range; IQR) from baseline (i.e. 1 January 2012) to their first CMM was 2.8 years (1.4–3.9) (range 0–5.0 years). The median (IQR) age at CMM diagnosis was 67 years (57–75) (Table V).
Primary analysis
The AUC for correctly identifying individuals with CMM was 0.59 (95% CI 0.57–0.61). The point on the ROC where sensitivity equalled specificity had a value of 56% (sensitivity 95% CI 53–60% and specificity 95% CI 55–58%) (Fig. 2).
Sensitivity (post-hoc) analyses
The model that used only ICD-10- and ATC-codes, but not demographic data, had an AUC of 0.56 (95% CI 0.54–0.58). The models that censored the last 39 weeks did not perform significantly worse than the corresponding non-censored models (p = 0.98). Moreover, using 2 convolutional layers did not alter the performance compared with similar models with 1 layer (p = 0.79). The model that included only demographic data performed on par with the primary model that included ICD-10-, ATC-codes and demographic data (Table III).
DISCUSSION
In this pilot investigation, using only routinely sampled registry data, we were able to predict the risk of development of CMM within a 5-year time period with an AUC of 0.59, when matching individuals with respect to age and sex. Non time-dependent variables (including origin, marital status, education level, disposable income, and region of birth) played a more important role for the model performance compared with time-dependent variables (including ATC- and ICD-10 codes).
In a publication by Wang et al. (15), the authors conducted a retrospective analysis of a randomized set of the Taiwanese population. The aim was to generate a prediction model for 1-year risk of non-melanoma skin cancer (NMSC) in previously cancer-free individuals, based on routinely sampled healthcare registry data. Similar to our model, their CNN did not include traditional risk factors, such as sun exposure, smoking, or family history of skin cancer. Instead, the prediction model included sequential diagnoses and a selection of prescription codes for the past 3 years. The training of the model used 5-fold cross-validation, which included 1,829 individuals with NMSC and 7,665 randomly selected controls. The sensitivity (standard deviation; SD) and specificity (SD) for identifying those individuals who developed NMSC was 83.1% (3.5) and 82.3% (4.1), respectively. The network achieved an AUC of 0.89 (0.007) and a positive predictive value (SD) of 57.1% (4.9). The authors concluded that the predictive analytic model may help healthcare professionals to target high-risk populations and optimize prevention strategies. However, the control individuals were not matched with respect to age, meaning that they were significantly younger (47.5 years) than the corresponding individuals who had NMSC (65.3 years). In their investigation age alone was an important predictor for NMSC. Most importantly no holdout validation set nor external test set was used (16, 17). This limits the external validation of the findings to the general population at large. Finally, the investigation included a population with Asian descent, which limits the external generalizability to other populations.
This investigation has some important limitations. For this binary classification problem, a CNN model was employed; however, other approaches could have been used, including recurrent neural networks, random forests and gradient boosting (18). While the model outperformed chance alone, the overall AUC (0.59, 95% CI 0.57–0.61) for predicting CMM within 5 years, is low and far from acceptable to be used in routine healthcare. Nonetheless, we cannot rule out that other ML models would outperform a CNN in this setting. Future investigations with direct comparison of the performance level of a variety of ML architectures would be useful to investigate the most appropriate modality for this setting. The peak AUC, using registry data alone, that can be expected for CMM prediction is yet to be determined. Moreover, what AUC levels would be acceptable to move forward with this application as a tool in healthcare is also subject to debate. Even then the place for these algorithms must be clearly defined. One potential application would be to use this tool as a guide to select individuals for CMM screening or CMM prevention campaigns. Another, and perhaps more clinically feasible, application is to let trained physicians use the algorithmic output as an aid when weighing risk factors for skin cancer in the in- and out-patient settings. A complete patient history, including all previous diagnoses and a detailed list of all medications dispensed for the past decade, is of, course, impossible to systematically compile during a patient visit. However, if this data, including an algorithmic output of the future risk of skin cancer, were available using simple computer techniques it might add value to the physicians and could help enable personalized precision medicine. Moreover, if clinicians could gain access to the registry data that is important, the usability of this method would probably increase further. This type of application would only be suitable for physicians who have received adequate training in interpretation of algorithmic output. Finally, it should preferably be used when a physician is able to integrate other risk factors that are not captured in the registries.
This investigation included time-dependent variables (dispensed pharmaceutical drugs and diagnoses). This deserves particular mention as they, intrinsically, may indicate information about future events (i.e. disease evolution). However, we believe that this is of minor importance, since our post-hoc analysis, in which the last 39 weeks of time-dependent variables were censored, yielded results on par with our primary model. However, to limit this issue further all individuals with a history of CMM at 31 December 2011 were excluded. Notably, the model only including demographic data performed on par with our original model including demographic data, ATC- and ICD-10 codes. One possible explanation could be that the demographic data already captures the relevant information in the ICD-10- and ATC-codes needed to predict CMM. However, the models in which demographic data were omitted still performed better than chance alone (AUC, 0.56). While there are thousands of available ATC- and ICD-10 codes, in order to preserve computing memory, a generic lossless embedding was used, in which each character in the respective position in the code was simply given its own row (Table II). This means that if several different ICD-10 or ATC codes, respectively, occurred in the same week, then the model could not distinguish between them. However, the model could still learn that certain combinations of characters are probabilistically related to certain outcomes. In upcoming investigations, it is our intention to update the model and include a less compressed version of the set of these codes.
Importantly, our investigation was performed in a population that is comprised mainly of individuals with Fitzpatrick skin types ranging from I to III. Furthermore, melanoma incidence in Sweden is high (9), and Swedish citizens have universal access to healthcare. The external validity of the current findings is, by design, intrinsically limited to the Swedish population alone, and the usefulness of similar prediction models in other populations with more limited access to healthcare might be more restrained. Finally, while 1.2 million individuals were used as eligible controls in the current investigation, also drawn from the general population, the controls represent a non-random subset of the corresponding Swedish adult population (approximately 6.7 million individuals). In future studies, the complete adult population will be used to further develop, refine, and update new registry-based prediction models for skin cancer including CMM. While computational phenotyping holds promise in this setting, future prospective clinical trials integrating algorithmic output with relevant clinical metadata will be required to fully assess the potential of these ancillary tools. Moreover, even if these models perform well in a research setting, political stakeholders and legislation must be involved before any broad implementation of these tools can be made in everyday clinical practice.
Research involving ML prediction models, based on registry data, is still in its initial stages, and adequate standardization is still pending (19). Nonetheless, the current investigation illustrates the potential usefulness of computational phenotyping for risk assessment in prediction of CMM in a Swedish population.
ACKNOWLEDGEMENTS
This study was financed by grants from the Swedish state under agreement between the Swedish Government and the county councils; the ALF-agreement (ALFGBG-965546), The Gothenburg Society of Medicine (Göteborgs Läkaresällskap) (grant number: 973007) and HudFonden (Reference Number: 3205/2021:1).
The authors have no conflicts of interest to declare.
REFERENCES
- Saba L, Biswas M, Kuppili V, Cuadrado Godia E, Suri HS, Edla DR, et al. The present and future of deep learning in radiology. Eur J Radiol 2019; 114: 14–24.
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–118.
- Haenssle HA, Fink C, Schneiderbauer R, Toberer F, Buhl T, Blum A, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol 2018; 29: 1836–1842.
- Han SS, Park GH, Lim W, Kim MS, Na JI, Park I, et al. Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: Automatic construction of onychomycosis datasets by region-based convolutional deep neural network. Plos One 2018; 13: e0191493.
- Chu YS, An HG, Oh BH, Yang S. Artificial intelligence in cutaneous oncology. Front Med (Lausanne) 2020; 7: 318.
- Wang S, Yang DM, Rong R, Zhan X, Xiao G. Pathology image analysis using segmentation deep learning algorithms. Am J Pathol 2019; 189: 1686–1698.
- Cheng Y, Wang F, Zhang P, Hu J. Risk prediction with electronic health records: a deep learning approach. Proceedings of the 2016 SIAM International Conference on Data Mining: SIAM, 2016: p. 432–440.
- Du AX, Emam S, Gniadecki R. Review of machine learning in predicting dermatological outcomes. Front Med (Lausanne) 2020; 7: 266.
- Whiteman DC, Green AC, Olsen CM. The growing burden of invasive melanoma: projections of incidence rates and numbers of new cases in six susceptible populations through 2031. J Invest Dermatol 2016; 136: 1161–1171.
- Buja A, Sartor G, Scioni M, Vecchiato A, Bolzan M, Rebba V, et al. Estimation of direct melanoma-related costs by disease stage and by phase of diagnosis and treatment according to clinical guidelines. Acta Derm Venereol 2018; 98: 218–224.
- Tinghog G, Carlsson P, Synnerstad I, Rosdahl I. Societal cost of skin cancer in Sweden in 2005. Acta Derm Venereol 2008; 88: 467–473.
- Giannopoulos F, Gillstedt M, Laskowski M, Bruun Kristensen K, Polesie S. Methotrexate use for patients with psoriasis and risk of cutaneous squamous cell carcinoma: a nested case-control study. Acta Derm Venereol 2021; 101: adv00365.
- Polesie S, Gillstedt M, Paoli J, Osmancevic A. Methotrexate treatment for patients with psoriasis and risk of cutaneous melanoma: a nested case-control study. Br J Dermatol 2020; 183: 684–691.
- Statistics Sweden, Population Statistics. [accessed 2021 Dec 9]. Available from: https://www.statistikdatabasen.scb.se/pxweb/en/ssd/START__BE__BE0101__BE0101A/BefolkningR1860N/.
- Wang HH, Wang YH, Liang CW, Li YC. Assessment of deep learning using nonimaging information and sequential medical records to develop a prediction model for nonmelanoma skin cancer. JAMA Dermatol 2019; 155: 1277–1283.
- Vivot A, Gregory J, Porcher R. Application of basic epidemiologic principles and electronic health records in a deep learning prediction model. JAMA Dermatol 2020; 156: 472–473.
- Cho SI, Lee D, Jo SJ. Application of basic epidemiologic principles and electronic health records in a deep learning prediction model. JAMA Dermatol 2020; 156: 473–474.
- Richter AN, Khoshgoftaar TM. Efficient learning from big data for cancer risk modeling: a case study with melanoma. Comput Biol Med 2019; 110: 29–39.
- Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet 2019; 393: 1577–1579.