The aim of this study was to investigate the early-life development of the skin microbiome in atopic dermatitis. Nineteen infants with atopic dermatitis and 19 healthy infants were evaluated 3 times, at 3 months intervals, within the first 30 months of life. Tape-strips were collected from volar forearms, cheeks, and eczema lesions, and the skin microbiome was assessed by 16S rRNA sequencing. Both the community structure and richness of the skin microbiome of infants with atopic dermatitis differed significantly from that of healthy infants, with greater richness in healthy infants. For infants with atopic dermatitis, the community composition was not dominated by Staphylococci. For healthy infants, community composition and richness correlated significantly with age, while such a pattern was not revealed in infants with atopic dermatitis. This suggests a slower maturation of the skin microbiome in atopic dermatitis, which precedes the staphylococcal predominance observed in older children and adults.
Key words: atopic dermatitis; skin microbiome; infant.
Accepted Jul 12, 2022; Epub ahead of print Jul 12, 2022
Acta Derm Venereol 2022; 102: adv00817.
DOI: 10.2340/actadv.v102.2275
Corr: Caroline M. Olesen, Department of Dermatology, Bispebjerg Hospital, University of Copenhagen, Bispebjerg Bakke 23, DK-2400 Copenhagen NV, Denmark. E-mail: caroline.meyer.olesen@regionh.dk
SIGNIFICANCE
This study investigated how bacteria on the skin of infants with atopic dermatitis develop over time compared with in healthy infants. Infants with atopic dermatitis had different types of skin bacteria and a smaller number of different bacterial species compared with healthy subjects. The bacterial communities in healthy skin developed in an age-related way, with increasing numbers of bacterial species. Such a pattern was not observed in infants with atopic dermatitis, suggesting an altered maturation of skin bacteria. Understanding the early-life dynamics of skin bacteria in atopic dermatitis may provide insight into the complex disease mechanism of this condition and lead to the development of new preventive strategies.
INTRODUCTION
Atopic dermatitis (AD) is a highly prevalent inflammatory skin disease, affecting 20% of children in Western countries (1). Both genetic and environmental factors contribute to disease development, and the pathophysiology of AD involves interplays between impaired skin barrier function and activation of Th2 polarized immune responses (2, 3). Changes in the skin microbiome may contribute to disease pathogenesis, and the skin microflora in AD has been shown to be dominated by Staphylococcus aureus and decreased bacterial α-diversity, with these changes correlating with disease severity (4–6). Mechanistic studies have revealed a mutually reinforcing relationship between S. aureus and AD skin conditions, in which S. aureus is both promoted by conditions in AD skin and aggravates the disease (7, 8). Accordingly, the skin microflora has been hypothesized to play a role in the development and worsening of AD, potentially implicating a target for therapies and prevention (9).
Increasing evidence suggests that microbial communities play a pivotal role in shaping the host immune system. Microbial imbalance has thus been implicated in the development of multiple diseases, including AD (10–12). While most research has focused on the gut microbiome, the role of the skin microbiome has recently gained increased attention. However, at present, only a few studies have investigated the skin microbiome in early life AD (13, 14), showing decreased abundance of certain commensal staphylococci (14) and increased colonization with S. aureus (15) prior to the development of AD. These data suggest that dysregulation in the early skin microbiome may promote AD pathology. However, further insights in the dynamic state of the infantile skin microbiome in healthy and AD subjects are needed to explore the potential of the skin microbiome as a target for preventive strategies.
The aim of this study was to investigate whether the temporal development of the bacterial communities in infants with AD differs from that in healthy controls.
MATERIALS AND METHODS
Participants
A total of 19 infants with AD and 19 healthy infants were included in the study. The infants with AD were recruited through the outpatient clinic at the Department of Dermatology, Bispebjerg Hospital. Healthy infants were recruited through local advertising. All participants were included from November 2017 to July 2018. For infants with AD, inclusion criteria comprised a diagnosis of AD according to UK criteria (16), age below 2 years at inclusion, and no treatment with systemic immunosuppressive drugs. Inclusion criteria for healthy infants comprised age below 2 years and no treatment with pharmaceutical drugs.
Study design and skin sampling
Participants were evaluated 3 times, at inclusion and at 3- and 6-month follow-ups. At each evaluation, disease severity was assessed with objective SCORing Atopic Dermatitis (O-SCORAD) and skin samples were collected by D-squame tape-strips (CuDerm, Dallas, Texas, USA). For all infants, 1 tape-strip were collected from the cheek and one from the volar forearm (VF). For the AD infants, 1 tape-strip was also collected from a skin lesion (Fig. 1).

DNA extraction and library preparation
Swabs were extracted as per Barnes et al. (17), using a modified protocol and the Blood and Tissue Kit (QIAGEN, Copenhagen, Denmark). Similarly, sequencing preparation was also outlined in Barnes et al. (2020) using the tagged V3-V4 1S rRNA primers 341F (5’-CCTAYGGGRBGCASCAG-3’) and 806R (5’-GGACTACNNGGGTATCTAAT-3’). PCR negatives were run with every batch of PCRs (approximately 1 every 32 reactions) and extraction negatives were included. Samples and blanks were split randomly amongst 3 runs on the Illumina 4000 MiSeq platform (Illumina, San Diego, CA, USA) within the National High-Throughput Sequencing Centre, University of Copenhagen, Denmark.
Bioinformatics
Libraries were demultiplexed using internal tags attached to primers, and they underwent processing as per Barnes et al. (17), using DADA2 (18). However, processed reads were normalized using a fourth-root transformation instead of using DeSeq2, before finally being converted to relative abundances. In total, 25 negative controls were run, and amplicon sequence variants (ASVs) were removed if found in more than 7 negative controls. In addition, samples with a low abundance of reads (< 2,000 reads) were removed. Therefore, from an initial 29,669,097 unpaired reads that were assigned to samples, 11,504,969 paired reads (mean of 37,597 reads per sample) were retained after quality filtering, removal of samples with a low number of reads, and removal of possible contaminant ASVs.
Statistical analysis
All statistical analyses were performed within the statistical computer language R and visualized using the gplot2 package within it. Jaccard and Yue-Clayton similarity values were both calculated from custom scripts (https://github.com/drcjbarnes/AD_within_infants).
Initially, inter-individual variation was analysed against the community composition using PERMANOVA (adonis function of vegan) and ASV richness using generalized linear modelling, with significance testing performed using likelihood-ratio testing. Given the large effects of inter-individual variation, community composition and ASV richness were correlated against sampling visit (visit number 1, 2 and 3), skin site (AD-lesional, AD-VF, AD-Cheek, Healthy control (HC)-VF, HC-Cheek) and age (in months) while controlling for inter-individual variation using PERMANOVA and mixed linear modelling (MLM; significance testing also performed using likelihood-ratio testing).
Lesional samples were removed to create like-for-like comparisons between patients with AD and healthy controls (i.e. volar forearms and cheek samples from both). The effects of AD status and anatomical location (volar forearm, cheek) were assessed using PERMANOVA for community composition and MLM for ASV richness (controlling for inter-individual variation). Similarly, AD samples were analysed without healthy controls, with sampling visit, age, anatomical location and severity (measured as O-SCORAD) correlated against the community composition and ASV richness, also using PERMANOVA and MLM. The bacterial communities for each body location for lesional and non-lesional (AD-Cheek and AD-VF) skin were also analysed separately, using PERMANOVA and MLM to correlate community variation and ASV richness against visit, age, O-SCORAD (AD samples only) and the anatomical location of the lesion.
The specific taxa associated with age were explored within the healthy control samples. Major bacterial families were identified if they had a mean relative abundance above 3% or mean ASV richness of 5 (and occurring in more than 3 samples). These then underwent mixed linear modelling and likelihood ratio testing as before, using patient number as the random effect. Similarly, major individual ASVs were identified (mean relative abundance of more than 0.1%) and correlated against age, using MLM. The specific families and ASV associated with AD were also explored. For the non-lesional samples, MLM was performed as for the healthy controls, substituting age for O-SCORAD. The relative abundance and ASV richness of bacterial families, and abundance of major individual ASVs within the lesional samples were also correlated against O-SCORAD, which was performed using Spearman’s rank sum correlations.
Ethical considerations
The study was approved by the local ethics committee (Hillerød, Denmark, project ID: H-16047983) and by the Danish Data Protection Agency (BFH-2017-042, I-Suite: 05449). Oral and written information were provided by the investigator, and written informed consent was obtained.
Data availability statement
Sequencing data and supporting information were uploaded to the University of Copenhagen’s Electronic Research Data Archive (ERDA), where it is freely available for download (https://erda.ku.dk/archives/c0136a84eef1224a826712b56d2fc81c/published-archive.html).
RESULTS
Nineteen infants with AD (mean age at inclusion (range): 12.5 months (5–22); female: 31.6%; mean O-SCORAD (range): 19.0 (8.0–31.9)) and 19 healthy infants (mean age at inclusion (range): 8.5 months (3–21); female: 57.9%) were included in the study (Table SI). Patients were allowed to use topical treatments (i.e. corticosteroids and calcineurin inhibitors) on the skin lesions. A total of 5 infants with AD were treated with antibiotics within 14 days prior to sampling (topical: 3, systemic: 2) while none of the healthy infants were treated with antibiotics within 14 days of sampling.
Overall community composition across all samples
The bacterial composition was in accordance with previous studies of infants (10), with the Bacteroidales (mean relative abundance of 13.98%, mean number of ASVs 8.96), Burkholderiales (10.55%, 8.55 ASVs) and Lactobacillales (10.21%, 7.28 ASVs) being the most dominant orders across all samples, and Streptococcus (5.1%, 5.3 ASVs), Corynebacterium (3.8%, 5.7 ASVs) and Veillonella (3.1%, 3.6 ASVs) being the most abundant at genus level.
The community composition was correlated against inter-individual variation (i.e. participant ID), skin site (a combination of skin status (i.e. AD vs healthy skin) and anatomical location (AD-lesion, AD-VF, AD-Cheek, HC-VF, HC-Cheek)), age (in months), and sampling visit (visits 1, 2 and 3)). Age refers to the impact of biological age on the community composition (i.e. are a 20-month-old infant and a 21-month-old infant more similar in community composition than a 3-month-old and a 21-month-old) and correlations with age suggest predictable variation in the bacterial communities of infants from birth to 30 months of age. Sampling visit refers to community similarity over time within individuals (i.e. are the communities more alike from visits 1 and 2 than visits 1 and 3).
Inter-individual variation had the strongest correlation with the community composition, accounting for 26.1% of community variation. Community composition likewise correlated with temporal variation (i.e. age and visit), with age accounting for 0.9% and sampling visit for 1.0% of community variation (Table I). There was no correlation between anatomical location and community composition. ASV richness correlated only with inter-individual variation and none of the other factors (Table I).

The impact of AD status was explored by removing lesional samples, allowing for direct paired comparisons between AD non-lesional skin and healthy control skin. Both the community composition (R2=0.008, p = 0.001) and ASV richness (LRT 7.341, p = 0.007) were significantly different between AD non-lesional and healthy control skin, with lower ASV richness in AD infants (139.7 ASVs) compared with healthy controls (187.7 ASVs) (Fig. 1A).
Skin microbiome of infants with atopic dermatitis did not vary with age contrary to healthy infants
Data was partitioned into individual skin sites (AD-Lesion, AD-VF, AD-Cheek, healthy control HC-VF, HC-cheek) and differences in the bacterial communities associated with sampling visit and age were explored (Table II, Fig. 2). Within the HC samples, significant temporal variation in the community composition of both cheeks and VF was observed (Fig. 2C), with significant shifts in community composition with both sampling visit and age. ASV richness also significantly correlated with both sampling visit and age within the HC-VF samples (Fig. 2B, Table II), showing increase in richness over time (approximately 175 ASVs at 3 months of age to 250 ASVs at 24 months of age). Within AD samples, there were significant differences in the composition of the AD-VF communities between sampling visits, while neither community composition nor ASV richness correlated with sampling visit in the AD-Cheeks and AD-Lesion samples. Age did not correlate with either community composition or richness in any of the AD skin sites (Table II).
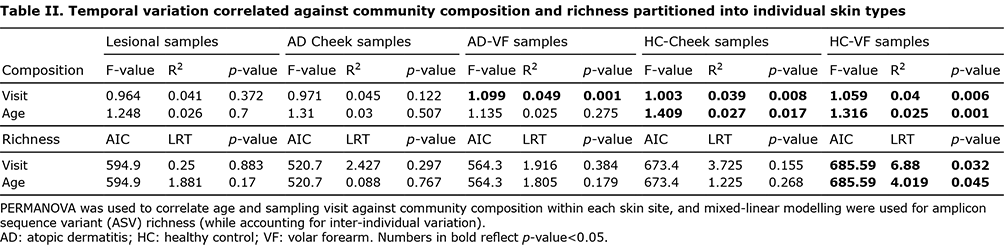
The bacterial families that varied with age were investigated within the healthy controls. There were significant increases in the ASV richness of the Corynebacteriaceae and Micrococcaceae families and significant decreases in the abundances of the Streptococcaceae and Veillonellaceae.
Analyses within patients with atopic dermatitis
AD severity (measured as O-SCORAD) was correlated against the community composition and ASV richness of infants with AD. The AD samples analysed together (AD-Lesion, AD-VF and AD-cheek) showed a significant negative correlation between AD severity and ASV richness (LRT 1.881 p = 0.017), but did not correlate with the community composition (R2=0.019, p = 0.248) (Fig. S1). However, there were no correlations when the individual skin types were analysed separately. The effect of changing O-SCORAD was further explored within specific bacterial families. Family abundance and ASV richness were correlated against O-SCORAD, revealing no significant correlations.
Bacterial communities of the older infants more closely represented adults
As part of Barnes et al. (19) study, 20 adults with AD and 20 healthy controls were sampled repeatedly at the same clinic during the same period of time, using tape-stripping. To investigate the similarities between infants and adults, the mean composition for each skin group was calculated at different age ranges (0–6, 7–12, 13–18, 18+ months, and adults) and compared at the family level (Fig. 3). While the community composition was similar throughout life (from infancy to adult), there was an increase in ASV richness from a broad range of families in the adults. Likewise, there was an increased ASV richness from a range of families observed in the 18+ healthy controls, which was not observed in AD infants (Fig. 3).

DISCUSSION
This study investigated the temporal changes in the skin microbiome of infants with AD and in healthy infants. While the bacterial communities of both AD and healthy skin underwent significant temporal variation between sample time-points, only healthy skin showed age-related changes in community composition and increasing bacterial richness towards an adult community assemblage. Thus, these results suggest a slower maturation of the skin microbiome in infants with AD than in healthy subjects.
Previous studies have shown that the adult microbiome of healthy skin is dominated by Proteobacteria, Actinobacteria and Firmicutes (20, 21), and that the microbiome of healthy infants shows increased α-diversity with age (22). Thus, the current results for healthy infants are in line with these previous studies and suggest that the maturation of the AD skin microbiome is altered. It can be speculated whether this change in maturation of the skin microbiome in AD may be related to establishment of the characteristic dysregulation of the microbiome in older children and adults with AD with decreased α-diversity and increased abundance of S. aureus (23–25).
In accordance with previous studies (9, 26), the microbiome of non-lesional AD skin displayed decreased richness and significantly different community composition compared with healthy skin. However, contrary to findings in adults, the AD skin microbiome was not dominated by Staphylococci. This could indicate that the reduced α-diversity (i.e. richness and evenness) in infants with AD is not primarily driven by S. aureus predominance, as seen in older children (5, 27). Furthermore, it can be speculated whether these early-life alterations of the AD skin microbiome drive the microbiome towards decreased richness, creating favourable conditions for subsequent S. aureus colonization.
The lack of staphylococcal predominance revealed in the current study is in accordance with the findings of a microbiome study by Kennedy et al. (14), showing decreased commensal Staphylococci prior to AD development, but no increases in S. aureus abundance in infants developing AD compared with healthy subjects. However, a study by Meylan et al. (15) did find increased S. aureus colonization prior to AD development by culture-based methods, possibly reflecting differences in methodology.
Surprisingly, the current study found a limited relationship between AD severity (i.e. O-SCORAD) and the skin microbiome, although there was a significant decline in ASV richness associated with increasing O-SCORAD. This is inconsistent with findings in older children and adults (4, 5, 28, 29). Here, SCORAD was mild to moderate at inclusion and improved during the follow-up period, and ultimately may not have been severe enough to drive major shifts in the skin microbiome. However, this may also suggest that the correlation between the skin microbiome in AD and severity is established later in life. This finding further emphasizes that severity is not driving the observed difference between AD and healthy infants, suggesting internal host factors (e.g. altered expression of immunological mediators) as key drivers of this difference.
Study limitations
This study has some limitations, including the relatively small number of participants. Inclusion of a larger number of participants may have led to identification of age-related taxonomical shifts that differed between AD and healthy individuals. Furthermore, variation of the anatomical location of lesional sample sites and topical treatment of the lesions may have influenced the results on the lesional skin microbiome, potentially explaining the lack of correlation with disease severity. However, we accounted for this by removing the lesions for some of the analyses, thereby revealing differences between AD non-lesional and healthy skin. Mean age of the healthy infants were 4 months younger than infants with AD, potentially influencing results on age-dependent changes. However, when stratified into age groups, the age-related pattern was observed across all age groups and not just in the youngest groups.
Conclusion
This study showed a different temporal development of the skin microbiome in both lesional and non-lesional skin of infants with AD compared with healthy subjects, suggesting a slower maturation of the AD skin microbiome towards an adult community assemblage. Contrary to findings in adult AD, decreased richness and altered community composition of the AD skin microbiome was not driven by a predominance of Staphylococci. Further studies of young children beyond the initial years of life are required to establish at which age a full adult microbiome is reached, and whether it is delayed in AD. Furthermore, the significance of these early-life microbiome alterations in shaping the cutaneous immune system should be explored in order to establish the potential clinical implications of these findings with respect to prevention.
ACKNOWLEDGEMENT
This work was funded by the Leo Pharma Foundation.
The authors have no conflicts of interest to declare.
REFERENCES
- Langan SM, Irvine AD, Weidinger S. Atopic dermatitis. Lancet 2020; 396: 345–360.
- Guttman-Yassky E, Waldman A, Ahluwalia J, Ong PY, Eichenfield LF. Atopic dermatitis: pathogenesis. Semin Cutan Med Surg 2017; 36: 100–103.
- Weidinger S, Beck LA, Bieber T, Kabashima K, Irvine AD. Atopic dermatitis. Nature Rev Dis Primers 2018; 4: 1.
- Clausen ML, Agner T, Lilje B, Edslev SM, Johannesen TB, Andersen PS. Association of disease severity with skin microbiome and filaggrin gene mutations in adult atopic dermatitis. JAMA Dermatol 2018; 154: 293–300.
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 2012; 22: 850–859.
- Totté JE, van der Feltz WT, Hennekam M, van Belkum A, van Zuuren EJ, Pasmans SG. Prevalence and odds of staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol 2016; 175: 687–695.
- Williams MR, Nakatsuji T, Gallo RL. Staphylococcus aureus: master manipulator of the skin. Cell Host Microbe 2017; 22: 579–581.
- Iwamoto K, Moriwaki M, Niitsu Y, Saino M, Takahagi S, Hisatsune J, et al. Staphylococcus aureus from atopic dermatitis skin alters cytokine production triggered by monocyte-derived Langerhans cell. J Dermatol Sci 2017; 88: 271–279.
- Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol 2019; 143: 26–35.
- Schoch JJ, Monir RL, Satcher KG, Harris J, Triplett E, Neu J. The infantile cutaneous microbiome: a review. Pediatr Dermatol 2019; 36: 574–580.
- Arora SK, Dewan P, Gupta P. Microbiome: paediatricians’ perspective. Indian J Med Res 2015; 142: 515–524.
- Casterline BW, Paller AS. Early development of the skin microbiome: therapeutic opportunities. Pediatr Res 2021; 90: 731–737.
- Zheng Y, Wang Q, Ma L, Chen Y, Gao Y, Zhang G, et al. Alterations in the skin microbiome are associated with disease severity and treatment in the perioral zone of the skin of infants with atopic dermatitis. Eur J Clin Microbiol Infect Dis 2019; 38: 1677–1685.
- Kennedy EA, Connolly J, Hourihane JO, Fallon PG, McLean WH, Murray D, et al. Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol 2017; 139: 166–172.
- Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, et al. Skin colonization by Staphylococcus aureus precedes the clinical diagnosis of atopic dermatitis in infancy. J Invest Dermatol 2017; 137: 2497–2504.
- Williams HC, Burney PG, Strachan D, Hay RJ. The U.K. Working Party’s Diagnostic Criteria for Atopic Dermatitis. II. Observer variation of clinical diagnosis and signs of atopic dermatitis. Br J Dermatol 1994; 131: 397–405.
- Barnes CJ, Rasmussen L, Asplund M, Knudsen SW, Clausen ML, Agner T, et al. Comparing DADA2 and OTU clustering approaches in studying the bacterial communities of atopic dermatitis. J Med Microbiol 2020; 69: 1293–1302.
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 2016; 13: 581–583.
- Barnes CJ, Clausen ML, Asplund M, Rasmussen L, Olesen CM, Yüsel YT, et al. Temporal and spatial variation of the skin-associated bacteria from healthy participants and atopic dermatitis patients. mSphere 2022; 7: e0091721.
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science 2009; 324: 1190–1192.
- Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol 2011; 9: 244–253.
- Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol 2011; 131: 2026–2032.
- Bjerre RD, Bandier J, Skov L, Engstrand L, Johansen JD. The role of the skin microbiome in atopic dermatitis: a systematic review. Br J Dermatol 2017; 177: 1272–1278.
- Kwon S, Choi JY, Shin JW, Huh CH, Park KC, Du MH, et al. Changes in lesional and non-lesional skin microbiome during treatment of atopic dermatitis. Acta Derm Venereol 2019; 99: 284–290.
- Olesen CM, Ingham AC, Thomsen SF, Clausen ML, Andersen PS, Edslev SM, et al. Changes in skin and nasal microbiome and staphylococcal species following treatment of atopic dermatitis with dupilumab. Microorganisms 2021; 9: 1487.
- Chng KR, Tay AS, Li C, Ng AH, Wang J, Suri BK, et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nature Microbiol 2016; 1: 16106.
- Seite S, Flores GE, Henley JB, Martin R, Zelenkova H, Aguilar L, et al. Microbiome of affected and unaffected skin of patients with atopic dermatitis before and after emollient treatment. J Drugs Dermatol 2014; 13: 1365–1372.
- Gonzalez ME, Schaffer JV, Orlow SJ, Gao Z, Li H, Alekseyenko AV, et al. Cutaneous microbiome effects of fluticasone propionate cream and adjunctive bleach baths in childhood atopic dermatitis. J Am Acad Dermatol 2016; 75: 481–493.e8.
- Callewaert C, Nakatsuji T, Knight R, Kosciolek T, Vrbanac A, Kotol P, et al. IL-4Rα Blockade by dupilumab decreases staphylococcus aureus colonization and increases microbial diversity in atopic dermatitis. J Invest Dermatol 2020; 140: 191–202.e7.