Tumour microenvironment has an important effect on the progression of cutaneous T-cell lymphomas. Using PCR with sequence-specific primers, this study analysed single-nucleotide polymorphisms in the interleukin-17 genes of 150 patients with cutaneous T-cell lymphoma. GG homozygote rs8193036 A/G of interleukin-17A gene occurred less commonly in the cutaneous T-cell lymphoma group; however, patients with this single-nucleotide polymorphism experience significantly intense pruritus. Conversely, the rs2397084 AG heterozygote of interleukin-17F is more common in the lymphoma population. In addition, there were significant differences in the frequencies of interleukin-17 genotypes when comparing early (Ia to IIa) and advanced stages (IIb, III and IV) of this neoplasms. A similar result has been shown in comparison between Sézary syndrome and mycosis fungoides. The current data may serve as a possible explanation for the increased bacterial infection rates in the course of cutaneous T-cell lymphoma, especially caused by Staphylococcus aureus. In summary, specific single-nucleotide polymorphisms occur with different frequencies between cutaneous T-cell lymphoma and healthy patients. Moreover, genetic predisposition of several interleukin-17 single-nucleotide polymorphisms may be a factor causing impaired immune defence in cutaneous lymphomas.
Key words: cutaneous lymphoma; mycosis fungoides; Sézary syndrome; cytokine; interleukin-17; lymphoma pathogenesis; single-nucleotide polymorphism.
Accepted Aug 16, 2022; Epub ahead of print Aug 16, 2022
Acta Derm Venereol 2022; 102: adv00777.
DOI: 10.2340/actadv.v102.2416
Corr: Karol Kołkowski, Dermatological Students Scientific Association, Department of Dermatology, Venerology and Allergology, Faculty of Medicine, Medical University of Gdansk, 17th Str. Smoluchowskiego, PL-80-214 Gdansk, Poland. E-mail: karolkolkowski@gumed.edu.pl
SIGNIFICANCE
Cutaneous T-cell lymphomas are a rare entity of lymphoproliferative disorders. This study presents genetic predisposition of some single-nucleotide polymorphic variants in the context of developing cutaneous T-cell lymphomas. Moreover, the results show some single-nucleotide polymorphisms of interleukin-17A and interleukin-17F to be significantly associated with pruritus intensity. These results may explain the increased susceptibility and rates of skin infections in the course of cutaneous T-cell lymphomas. Analysis of the current data and the literature may lead to improved care and management of patients with cutaneous T-cell lymphomas.
INTRODUCTION
Cutaneous T-cell lymphomas (CTCLs) belong to a group of rare, lymphoproliferative disorders, which have their origin in skin (1). Interleukin (IL)-17 has been implicated in the pathogenesis of these tumours, but the exact mechanism remains unclear (2–4). Various single-nucleotide polymorphisms (SNPs) of IL-17 genes have previously been associated with increased risk and worse course of autoimmune and neoplastic diseases (5–12). Other cytokines have been studied in this context in CTCL (13, 14). The aim of this study was to elucidate whether the frequency of selected SNPs of IL-17 genes are associated with susceptibility to the CTCL.
MATERIALS AND METHODS
The study was approved by the Independent Bioethics Committee for Scientific Research at Medical University of Gdańsk, Poland. A total of 150 blood samples of patients with CTCL: 139 MF in stages IA (44 cases), IB (38 cases), IIA (3 cases), IIB-IV (54 cases), and 11 Sézary syndrome (SS) diagnosed and treated at the Department of Dermatology of the Medical University in Gdansk and a control non-CTCL group of 196 unrelated healthy individuals within similar age and sex distribution, without personal or family history of chronic skin diseases, without pruritus and without personal history of malignancy were included in the study. Patients had been diagnosed on the basis of clinical, histopathological and immunohistochemical findings, according to the European Organization of Research and Treatment of Cancer (EORTC) criteria (1). Pruritus intensity was evaluated according to visual analogue scale (VAS) and numeric rating scale (NRS) and correlated with IL-17 gene polymorphisms.
DNA extraction/genotyping
Genomic DNA was isolated from all blood samples with the Blood Mini A&A Biotechnology (A&A Biotechnology, Gdansk, Poland) according to the instructions of the manufacturer. Analysis of the polymorphic variants IL-17A (rs2275913, rs3819024, rs8193036) and IL-17F (rs763780, rs2397084) were analysed by PCR with sequence-specific primers (SSP-PCR). Graphical visualization of this method is shown in Appendix S1.
Statistical analysis
Statistical calculations were made with Statistica, version 12.0 (StatSoft, Inc. 2015, Tulsa, Oklahoma, USA). Analysis of qualitative features was made with the χ2 test in the Pearson method. Independent variables fulfilling the assumptions for parametric tests were analysed with the Student’s t-test. Independent variables that did not meet the parametric test assumptions were analysed with non-parametric tests (analysis of variance (ANOVA) equivalents): Mann–Whitney U test (comparison of 2 tests) or Kruskal–Wallis test (comparison of many samples). Odds ratios (ORs) with 95% confidence intervals (95% CI) were determined by a logistic regression. In all tests, p < 0.05 was considered a significant level of statistical significance.
RESULTS
Genotype distribution of the studied SNPs (AA, AG, and GG) of IL-17A and IL-17F in which significant differences between studied groups in the distribution of alleles were found are shown in Tables I–III.
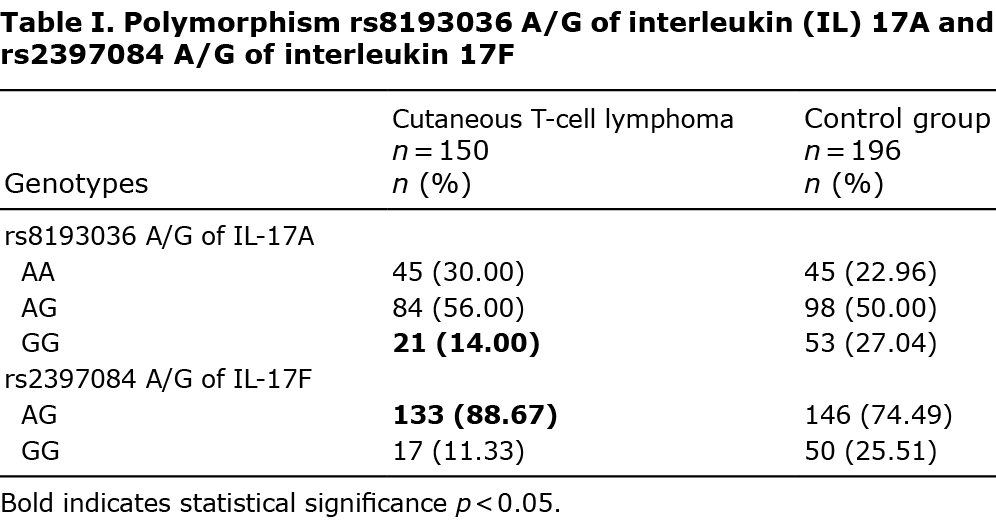
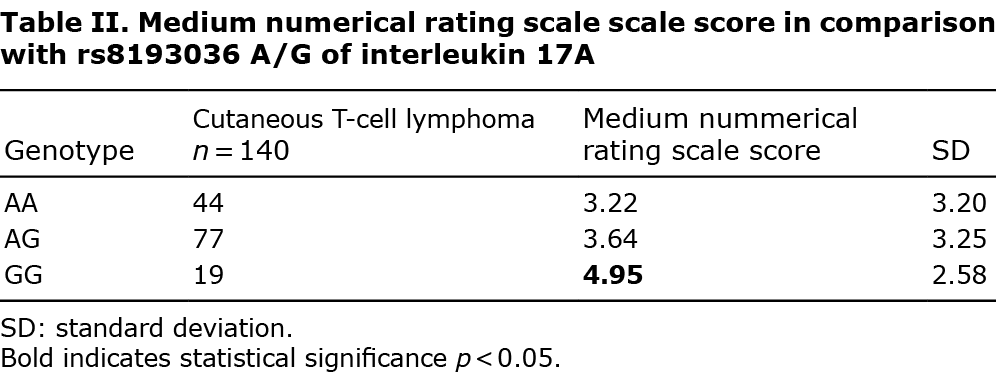
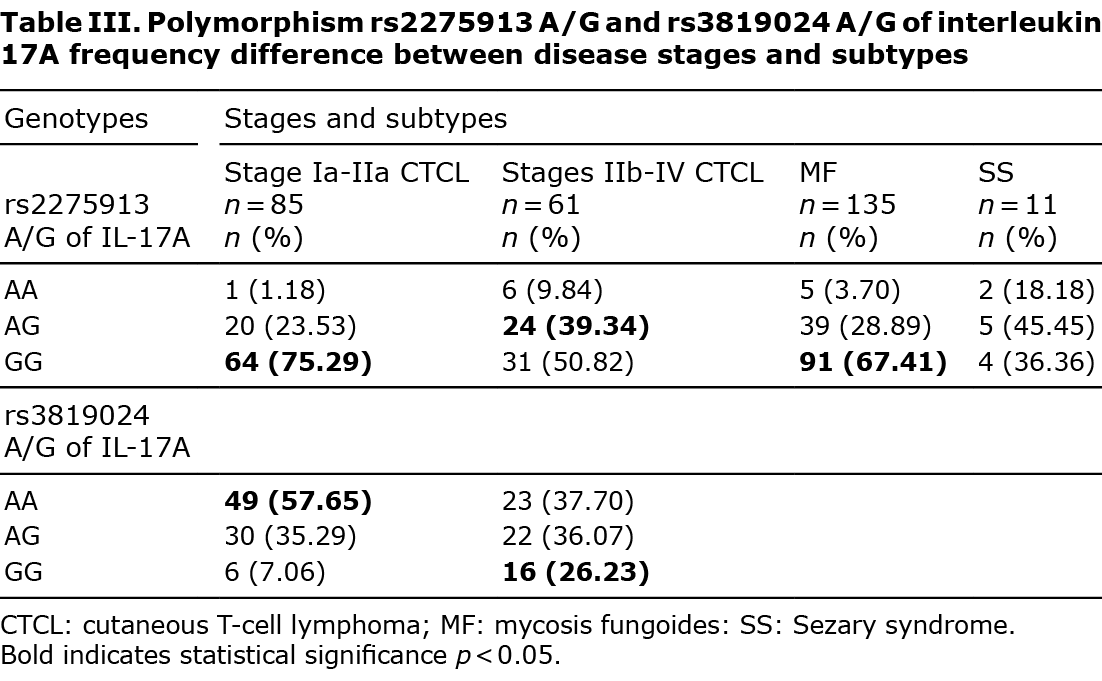
Interleukin-17A gene polymorphisms
Of the 3 studied loci, the only polymorphism which occurred significantly different in comparison with healthy controls was rs8193036 A/G of interleukin 17A. The presence of the IL-17A GG genotype has been found to be less frequent in the current CTCL population (odds ratio (OR) 0.4392, 95% CI 0.2512–0.7679 and p = 0.0039) (Table I). The pruritus intensity regarding our group has also been studied. Medium NRS in IL-17A rs8193036 polymorphism appeared to be significantly higher when the GG homozygote was present, in comparison with other genotypes (p = 0.03) (Table II). Moreover, the distinction between pruritus level GG and other polymorphisms has also been discovered to be significant (p = 0.005) (Appendix S1).
IL-17F gene polymorphisms
Of the 2 studied loci of the IL-17F gene, the only polymorphism that occurred significantly more commonly in comparison with the control group was rs2397084 AG (OR 2.6793, 95% CI 1.4729–4.8739 and p = 0.0009) (Table I). As before, it is also the only polymorphism of IL-17F in the current study, in which the visual analogue scale (VAS) pruritus levels have differed significantly between AG and GG (p = 0.01) (Appendix S1).
Stage of a lymphoma and IL-17 polymorphisms
Several distinctions were found between the distribution of certain genotypes in the various stages of the CTCL. The distribution of polymorphisms in the early stage (Ia to IIa) and advanced (IIb and higher) stages of lymphomas were analysed. Presence of IL-17A AG and GG genotypes of the rs2275913 polymorphism were respectively more (OR 2.1081, 95% CI 1.0285–4.3210 and p = 0.0417) and less (OR 0.3391, 95% CI 0.1678–0.6852 and p = 0.0026) frequent in IIb, III and IV stage CTCL compared with early stages (Table III). The analysis was significant, although not reliable due to the small group size with the AA genotype. Similar observations have been made concerning rs3819024 A/G polymorphism of interleukin 17A. The AA genotype was less frequent in advanced CTCL (OR 0.4447, 95% CI 0.2268–0.8718 and p = 0.0183), while the GG homozygote was more common in the mentioned group (OR 4.6815, 95% CI 1.7099–12.817 and p = 0.0027) (Table III). Significant differences were found in the frequency of polymorphism rs2275913 A/G of interleukin 17A genotypes when comparing patients with SS with those with mycosis fungoides (MF). The GG homozygote was found more often in MF than in SS (OR 0.2763, 95% CI 0.0768–0.9939 and p = 0.0489) (Table III). No significant distinctions were found when analysing other polymorphisms in that manner (Appendix S1).
DISCUSSION
To our knowledge this is the first study of IL-17 SNPs in primary CTCL. Several SNPs of IL-17 genes were associated with susceptibility to CTCL. The GG homozygote of rs8193036 A/G of interleukin 17A occurred less often in the CTCL group (Table I); however, patients with this polymorphism experienced significant pruritus (Table II). The rs2397084 AG heterozygote of IL-17F was more common in the CTCL population (Table I). Moreover, statistically significant differences between rs2275913 A/G and rs3819024 A/G of interleukin 17A have also appeared when comparing early and advanced CTCLs (Table III). That result may explain the more rapid progression to advanced stages of CTCLs in some patients. It is well known that several patients present stable disease for years. GG homozygote in rs2275913 A/G of interleukin 17A has been more common in the group of non-SS patients (Table III). It may suggest that this genotype has a lower predisposition to develop this aggressive, fast-spreading, leukaemic cutaneous lymphoma.
The role of IL-17 in CTCL is complex and not fully understood. It has been reviewed recently in the context of introducing new agents, such as bimekizumab, brodalumab, ixekizumab and secukinumab, in the treatment of psoriasis (15). IL-17A and IL-17F belong to the IL-17 family and are thought to have pro-inflammatory activity (16). They have been important in promoting the answer against extracellular bacteria and fungi (16). The IL-17A and IL-17F SNPs have been previously proven to increase the risk of asthma and rheumatoid arthritis (9–11). Similar correlation has been found in the Spanish cohort of psoriatic patients, but not in the Polish population (8, 12). SNP may play a role as a prognostic factor (6). Moreover, a link between IL-17 functions and carcinogenesis has been implicated in several different neoplasms; for example, in colorectal cancer, but also in the non-melanoma skin cancers (17, 18). Furthermore, several SNPs of IL-17A and IL-17F have been shown to be significantly associated with higher risk of gastric, lung and cervical cancer (5, 7, 19). SNPs have been studied in the context of cutaneous lymphomas, showing that some variants of IL-2 and IL-13 could be estimated as a risk factor, while IL-10 and tumour necrosis factor alpha (TNF-α) have a protective effect against developing CTCL (13).
The exact pathogenesis of CTCL is unknown despite extensive previous research (20). Interestingly, recent studies of the mouse model have shown that T-cell receptor engagement is an important factor of malignant transformation in CTCL (20). Moreover, progression of the disease also appeared to be dependent on microbiota (20, 21). In addition, Staphylococcus aureus is known as the most common aetiological factor in infections in patients with CTCL, who are colonized by this bacteria in 44% up to 76% (22). S. aureus superantigens may exacerbate and/or perpetuate the clonal expansion of the lymphoma concomitantly with spreading cutaneous inflammation (21, 23). In addition, staphylococcal enterotoxin A isolates have been shown to induce signal transducer and activator 3 (STAT3) activation and expression of IL-17, which pathway has been hypothesized as one of the oncogenic factors in CTCL (3, 24). Recently, another link between staphylococcal enterotoxins and progression of the lymphoma has been noted: they have been shown to induce STAT5 and microRNA-155 (miR-155) (25). STAT3, STAT5 and miR-155 have been previously associated with CTCL pathogenesis (14). Cobomarsen, an inhibitor of miR-155 expression, is thought to be one of the promising novel therapies in CTCL, currently undergoing phase first-in-human clinical studies (26). The constant responses of both malignant and benign lymphocytes to the S. aureus inflammation may contribute to carcinogenesis, which would explain why transient, aggressive antibiotic therapy may slow the tumour progression in some cases (22, 27). S. aureus eradication may be a novel treatment for advanced MF/SS in the future (21, 28).
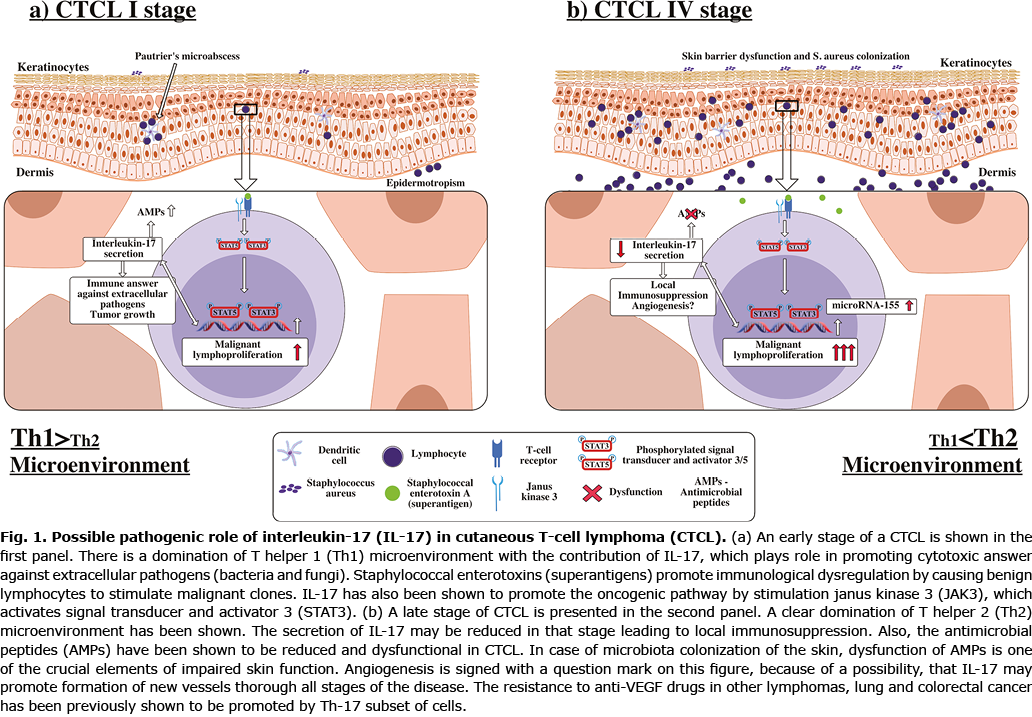
The disturbances in skin barrier also related to extensive inflammation in patients with lymphoma have been described previously (29, 30). Dysfunction of antimicrobial peptides secretion, which is induced by proinflammatory IL-17 cytokines, has been also observed (29, 30). The impaired skin function in case of infection can promote strong immune system defence. Despite elevated IL-17 levels in CTCL in several cases, such a state may lead to local immunosuppression caused by described mechanisms (2, 4, 24, 28, 31). We have summarized data on the possible pathogenic role of IL-17 in CTCL (Fig. 1). The results may provide a possible explanation for this phenomenon, as this study has revealed statistically significant differences in IL-17A rs8193036 GG and IL-17F rs2397084 AG SNPs in CTCL (Table I). In the case of IL-17A rs8193036 GG, more severe pruritus has been observed (Table II). Severe pruritus correlates with lower quality of life and both have been observed in advanced stages of MF and in SS (32). The statistical differences between early and advanced CTCL stages may also be an explanation of the fact that the pace of progression of the CTCL is individual. Bearing in mind that IL-17 belongs to a group of proinflammatory cytokines, and that Th2 profile is specific for advanced MF and SS, the rs2275913 A/G of interleukin 17A may protect switching from Th1 to Th2 profile in the CTCL microenvironment (Table III).
The study is limited by its design to test the specific SNPs instead of using new methods, such as next gene sequencing combined with the use of neural networks, which are currently shown in similar papers. It was not possible to perform them. However, the results of the current study seem to be important, especially in light of the pathogenetic influence of S. aureus in CTCL. As shown previously, blocking IL-17 by numerous new biologic drugs used in the therapy of, for example, psoriasis, may cause a progression and/or unmasking of the disease (15).
ACKNOWLEDGEMENTS
The authors thank Professor Jerzy Wojdylo from the Department of Mathematics, Southeast Missouri State University One University Plaza in Cape Girardeau, MO 63701, USA and Mrs Rae Wojdylo, who, as native speakers, have corrected the English language in this manuscript.
The study was financed by the Polish Ministry of Science and Higher Education grant. Project number ST-66. The study was supported by the Medical University of Gdańsk Project No. ST 02-10022 (0000701).
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by Independent Bioethics Committee for Scientific Research at Medical University of Gdańsk (decision number NKBBN/313/2017)
Informed consent was obtained from all subjects involved in the study.
The data presented in this study are available in Appendix S1.
The authors have no conflicts of interest to declare.
REFERENCES
- Willemze R, Cerroni L, Kempf W, Berti E, Facchetti F, Swerdlow SH, et al. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood 2019; 133: 1703–1714.
- Krejsgaard T, Litvinov IV, Wang Y, Xia L, Willerslev-Olsen A, Koralov SB, et al. Elucidating the role of interleukin-17F in cutaneous T-cell lymphoma. Blood 2013; 122: 943–950.
- Willerslev-Olsen A, Krejsgaard T, Lindahl LM, Litvinov I V, Fredholm S, Petersen DL, et al. Staphylococcal enterotoxin A (SEA) stimulates STAT3 activation and IL-17 expression in cutaneous T-cell lymphoma. Blood 2016; 127: 1287–1296.
- Miyagaki T, Sugaya M, Suga H, Kamata M, Ohmatsu H, Fujita H, et al. IL-22, but not IL-17, dominant environment in cutaneous T-cell lymphoma. Clin Cancer Res 2011; 17: 7529–7538.
- Omrane I, Marrakchi R, Baroudi O, Mezlini A, Ayari H, Medimegh I, et al. Significant association between interleukin-17A polymorphism and colorectal cancer. Tumor Biol 2014; 35: 6627–6632.
- Klonowska J, Gleń J, Nowicki RJ, Trzeciak M. Combination of FLG mutations and SNPs of IL-17A and IL-19 influence on atopic dermatitis occurrence. Adv Dermatol Allergol 2022; 39: 200–208.
- He Y, Du Y, Wei S, Shi J, Mei Z, Qian L, et al. IL-17A and IL-17F single nucleotide polymorphisms associated with lung cancer in Chinese population. Clin Respir J 2017; 11: 230–242.
- Bialecka M, Ostasz R, Kurzawski M, Klimowicz A, Fabiañczyk H, Bojko P, et al. IL17A and IL17F gene polymorphism association with psoriasis risk and response to treatment in a Polish population. Dermatology 2017; 232: 592–596.
- Shen L, Zhang H, Yan T, Zhou G, Liu R. Association between interleukin 17A polymorphisms and susceptibility to rheumatoid arthritis in a Chinese population. Gene 2015; 566: 18–22.
- Marwa OS, Kalthoum T, Wajih K, Kamel H. Association of IL17A and IL17F genes with rheumatoid arthritis disease and the impact of genetic polymorphisms on response to treatment. Immunol Lett 2017; 183: 24–36.
- Zhu M, Wang T, Chen R, Wang C, Liu S, Ji Y. Association between interleukin-17a gene polymorphisms and asthma risk: a meta-analysis. Asian Pacific J Allergy Immunol 2016; 34: 115–123.
- Batalla A, Coto E, González-Lara L, González-Fernández D, Gómez J, Aranguren TF, et al. Association between single nucleotide polymorphisms IL17RA rs4819554 and IL17E rs79877597 and psoriasis in a Spanish cohort. J Dermatol Sci 2015; 80: 111–115.
- Małgorzata SW, Nedoszytko B, Olszewska B, Roszkiewicz J, Glen J, Zabłotna M, et al. The role of polymorphism of interleukin-2, -10, -13 and TNF-α genes in cutaneous T-cell lymphoma pathogenesis. Adv Dermatology Allergol 2016; 33: 429–434.
- Olszewska B, Gleń J, Zabłotna M, Nowicki RJ, Sokołowska-Wojdyło M. The polymorphisms of IL-6/STAT3 signaling pathway may contribute to cutaneous T-cell lymphomas susceptibility. Arch Dermatol Res 2020; 313: 1–7.
- Kołkowski K, Sokołowska-Wojdyło M. Safety and danger of biologic treatments in psoriasis in context of cutaneous T-cell lymphoma (CTCL). Adv Dermatology Allergol 2021; 38: 953–960.
- Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol 2009; 9: 556–667.
- Wang K, Kim MK, DiCaro G, Wong J, Shalapour S, Wan J, et al. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity 2014; 41: 1052–1063.
- Nardinocchi L, Sonego G, Passarelli F, Avitabile S, Scarponi C, Failla CM, et al. Interleukin-17 and interleukin-22 promote tumor progression in human nonmelanoma skin cancer. Eur J Immunol 2015; 45: 922–931.
- Li L, Tian YL, Lv XM, Yu HF, Xie YY, Wang JD, et al. Association analysis of IL-17A and IL-17F polymorphisms in Chinese women with cervical cancer. Genet Mol Res 2015; 14: 12178–12183.
- Fanok MH, Sun A, Fogli LK, Narendran V, Eckstein M, Kannan K, et al. Role of dysregulated cytokine signaling and bacterial triggers in the pathogenesis of cutaneous T-cell lymphoma. J Invest Dermatol 2018; 138: 1116–11125.
- Licht P, Mailänder V. Transcriptional heterogeneity and the microbiome of cutaneous t-cell lymphoma. Cells 2022; 11: 328–331.
- Nguyen V, Huggins RH, Lertsburapa T, Bauer K, Rademaker A, Gerami P, et al. Cutaneous T-cell lymphoma and Staphylococcus aureus colonization. J Am Acad Dermatol 2008; 59: 949–952.
- Jackow CM, Cather JC, Hearne V, Asano AT, Musser JM, Duvic M. Association of erythrodermic cutaneous T-cell lymphoma, superantigen- positive Staphylococcus aureus, and oligoclonal T-cell receptor Vβ gene expansion. Blood 1997; 89: 32–40.
- Krejsgaard T, Ralfkiaer U, Clasen-Linde E, Eriksen KW, Kopp KL, Bonefeld CM, et al. Malignant cutaneous T-cell lymphoma cells express IL-17 utilizing the Jak3/stat3 signaling pathway. J Invest Dermatol 2011; 131: 1331–13338.
- Willerslev-Olsen A, Gjerdrum LMR, Lindahl LM, Buus TB, Pallesen EMH, Gluud M, et al. Staphylococcus aureus induces signal transducer and activator of transcription 5‒dependent miR-155 expression in cutaneous T-cell lymphoma. J Invest Dermatol 2021; 141: 2449–2458.
- Sugaya M. Clinical guidelines and new molecular targets for cutaneous lymphomas. Int J Mol Sci 2021; 22: 11079.
- Lindahl LM, Willerslev-Olsen A, Gjerdrum LMR, Nielsen PR, Blümel E, Rittig AH, et al. Antibiotics inhibit tumor and disease activity in cutaneous T-cell lymphoma. Blood 2019; 134:1072–1083.
- Fujii K. Pathogenesis of cutaneous T cell lymphoma: involvement of staphylococcus aureus. J Dermatol 2021; 49: 202–209.
- Suga H, Sugaya M, Miyagaki T, Ohmatsu H, Kawaguchi M, Takahashi N, et al. Skin barrier dysfunction and low antimicrobial peptide expression in cutaneous T-cell lymphoma. Clin Cancer Res 2014; 20: 4339–4348.
- Wolk K, Mitsui H, Witte K, Gellrich S, Gulati N, Humme D, et al. Deficient cutaneous antibacterial competence in cutaneous T-cell lymphomas: role of Th2-mediated biased Th17 function. Clin Cancer Res 2014; 20: 5507–5516.
- Cirée A, Michel L, Camilleri-Bröet S, Louis FJ, Oster M, Flageul B, et al. Expression and activity of IL-17 in cutaneous T-cell lymphomas (mycosis fungoides and Sezary syndrome). Int J Cancer 2004; 112: 113–120.
- Olszewska B, Żawrocki A, Gleń J, Lakomy J, Karczewska J, Zabłotna M, et al. Interleukin-31 is overexpressed in skin and serum in cutaneous T-cell lymphomas but does not correlate to pruritus. Adv Dermatology Allergol 2020; 39: 81–87.