ORIGINAL ARTICLE
Tumour Suppressor Neuron Navigator 3 and Matrix Metalloproteinase 14 are Co-expressed in Most Melanomas but Downregulated in Thick Tumours
Olga BUGAEVA1,2, Pilvi MALINIEMI1, Wenche S. PRESTVIK3, Eeva LEIVO1, Nicolas KLUGER1, Alexander SALAVA1, Sanna VIRTANEN4, Kirsi JÄNTTI1, Olli SAKSELA1, Kaisa LEHTI2,3, Paula KUJALA5, Kai KROHN4 and Annamari RANKI1
1Department of Dermatology and Allergology, University of Helsinki and Helsinki University Hospital, Helsinki, 2Research Program Unit, University of Helsinki, Helsinki, Finland, 3Department of Biomedical Laboratory Science, Norwegian University of Science and Technology, Trondheim, Norway and 4Clinical Research Institute HUCH, Helsinki, and 5Fimlab Laboratoriot Ltd, Tampere, Finland
Melanoma is a highly metastatic tumour originating from neural crest-derived melanocytes. The aim of this study was to analyse the expression of neuron navigator 3 (NAV3) in relation to membrane type-1 matrix metalloproteinase MMP14, a major regulator of invasion, in 40 primary melanomas, 15 benign naevi and 2 melanoma cell lines. NAV3 copy number changes were found in 18/27 (67%) primary melanomas, so that deletions dominated (16/27 of samples, 59%). NAV3 protein was found to be localized at the leading edge of migrating melanoma cells in vitro. Silencing of NAV3 reduced both melanoma cell migration in 2-dimensional conditions, as well as sprouting in 3-dimensional collagen I. NAV3 protein expression correlated with MMP14 in 26/37 (70%) primary melanomas. NAV3 and MMP14 were co-expressed in all tumours with Breslow thickness < 1 mm, in 11/23 of mid-thickness tumours (1–5 mm), but in only 1/6 samples of thick (> 5 mm) melanomas. Altogether, NAV3 number changes are frequent in melanomas, and NAV3 and MMP14, while expressed in all thin melanomas, are often downregulated in thicker tumours, suggesting that the lack of both NAV3 and MMP14 favours melanoma progression.
Key words: melanoma; naevus; NAV3; MMP14; copy number change.
SIGNIFICANCE
Melanoma is the most aggressive skin cancer, and its incidence is increasing. It is important to understand which genes and proteins are involved in the malignant transformation and progression of melanoma. This study found that tumour suppressor NAV3 was deleted in the majority of melanomas, and that NAV3 protein was expressed in all the thin melanomas, but was downregulated in thick tumours, suggesting that it plays a role in melanoma progression.
Citation: Acta Derm Venereol 2023; 103: adv00883. DOI https://doi.org/10.2340/actadv.v103.298.
Copyright: © Published by Medical Journals Sweden, on behalf of the Society for Publication of Acta Dermato-Venereologica. This is an Open Access article distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (https://creativecommons.org/licenses/by-nc/4.0/)
Accepted: Feb 6, 2023; Published: Mar 8, 2023
Corr: Olga Bugaeva, Department of Dermatology and Allergology, University of Helsinki and Helsinki University Hospital, Meilahdentie 2, FIN-00250 Helsinki, Finland. E-mail: Olga.Bugaeva@hus.fi
Competing interests and funding: The authors have no conflicts of interest to declare.
Finnish Cancer Foundation and Helsinki University Hospital Research Funds.
INTRODUCTION
Melanoma is one of the most feared cancers, since it is very therapy-resistant, and metastases can form even a decade after clinical remission. The incidence of melanoma is increasing worldwide, and the median age of patients is approximately 57 years, which is 10 years younger than that of all cancer patients (1). The prognosis of melanoma depends on its Breslow thickness and stage, as 5-year survival for stage I melanoma (localized) is 99% and for stage IV melanoma (with distant metastases) is 20% (2). Even though new treatments, such as immune checkpoint inhibitors and BRAF inhibitors, have markedly improved treatment results in patients with advanced melanoma, there is a lack of biomarkers that could predict which melanomas require sentinel lymph node biopsy and more aggressive treatment or predict the efficacy of new treatments.
Evolutionally conserved genes involved in cell proliferation and motility are likely to contribute to melanoma cell invasion and spreading. One such gene, neuron navigator 3 (NAV3), a human analogue to unc-3 (a cell guidance gene of C. elegans) belonging to the group of Navigator proteins that bind to the plus-ends of microtubules (+ TIPs, 3), has shown mutations, as well as copy number or expression changes in melanoma, glioblastomas, colorectal cancer, and several other benign and malignant types of tumours (4–14). In addition, NAV3 loss is associated with poor survival in breast cancer and nervous system tumours (15, 16). NAV3 inhibits tumour cell dissemination, most probably by promoting directional migration by stabilizing the microtubules and inhibiting random migration (16). Moreover, NAV3 is induced by tumour suppressor p73 (17), suggesting that it might regulate cancer metastasis.
The ability of tumour cells to modulate extracellular matrix is another prerequisite for invasion and metastasis. Matrix metalloproteinases (MMPs) are a large family of proteases that can degrade all components of extracellular matrix, cleave cell surface receptors, growth factors and other MMPs. Many of the MMPs are overexpressed in melanoma. For example, BRAF mutation leads to MMP9 overexpression, which induces melanoma cell invasion and neoangiogenesis (18). Membrane-bound matrix metalloproteinase 14 (MMP14, MT1-MMP) is often overexpressed in melanoma and regulates a plethora of cellular processes, including cell motility, extracellular matrix modulation, angiogenesis and metastatic spread (19). MMP2, which is activated by MMP14, also promotes melanoma spreading (20). The clear role of MMPs in tumour progression has led to the development of MMP inhibitors for cancer treatment. Although non-selective MMP inhibitors have failed in phase III studies due to lack of efficacy or adverse effects, new, more selective, treatment methods, such as MMP-responsive prodrugs, which are activated at the tumour site, are under intense investigation (21).
MMP14 is accumulated at the tips of invadopodia in the invading cancer cells, where it cleaves collagen type I and other matrix molecules enabling cancer cell invasion through interstitial tissues (22). Since microtubules are required for MMP14 trafficking to invadopodia (23), and NAV3 regulates microtubule formation, the aim of the current study was to explore the possible correlation of NAV3 and MMP14 expression in melanoma.
MATERIALS AND METHODS
Patient samples and melanoma cell lines
The study was approved by the Ethics Committee of Southern Finland (Dnr 492/E5/05 and 208/E5/01). Archival, paraffin-embedded tissue samples of primary melanomas (n = 27) and histologically benign naevi (n = 15), obtained for diagnostic purposes from patients treated at the Tampere University Central Hospital and Department of Dermatology and Allergology, Helsinki University Central Hospital, Helsinki, Finland, were analysed. In addition, the study examined another 13 melanomas (Breslow thickness ≥ 2.5 mm) for NAV3, MMP14 and MMP16 mRNA and protein expression. Two established melanoma cell lines, the primary WM-793 (VGP, vertical growth phase) cell line and the lymph node metastasis-derived WM-239 cell line provided by Dr Erkki Hölttä (Biomedicum Helsinki) and originally established by Dr Meenhard Herlyn (The Wistar Institute, Philadelphia, PA, USA (Wistar Institute melanoma cell line collection), were also used.
Probe labelling
Two bacterial artificial chromosome (BAC) clones specific to Nav3 DNA (RP11-36P3 and RP11-136F16; Research Genetics Inc., Huntsville, AL, USA) and the chromosome 12 centromere probe (pA12H8; American Type Cell Culture (ATCC), Manassas, VA, USA) were labelled with Alexa 594-5-dUTP (red; Thermo Fisher Scientific ,Waltham, MA, USA) and Alexa 488-5-dUTP (green; Thermo Fisher Scientific), respectively, using nick translation (5, 15). The probes were mixed with human COT-1 DNA (Invitrogen), precipitated and diluted into hybridization buffer (15% w/v dextran sulphate, 70% formamide in 2× Saline-Sodium citrate (SSC), pH 7.0).
Fluorescence in situ hybridization
For fluorescence in situ hybridization (FISH), cell nuclei were extracted, pretreated, and hybridized as described previously (5, 15, 24, 25). FISH signals were analysed both manually (by 2 blinded, independent researchers) and automatically, as described previously (24, 25). The manual analysis was performed using an Olympus BX61 microscope (Tokyo, Japan) equipped with a 60× and 100× oil-immersion objectives and a triple bandpass filter for simultaneous detection of Atto488, Alexa594 and DAPI (Chroma Technology Corp., Brattleboro, VT, USA) according to standard operating procedure (SOP Q2QC005; Dermagene Oy, Helsinki, Finland), which standardizes the signal scoring practice. The results obtained by automat (Metasystems Metafer4 software, Altlussheim, Germany) were checked manually by the 2 of the authors experienced in FISH analysis (PM, SV) and false results were deleted. The result was determined as false, if manually detected signals in the image of a single nucleus did not match with the results provided by the automat. A total of 200–1,000 nuclei were analysed from each case and the nuclei were grouped as normal if they had 2 signals for chromosome 12 centromere and 2 for the NAV3. Relative NAV3 deletion was defined when the number of NAV3 signals was lower than the number of centromere signals and relative NAV3 amplification was defined when the number of NAV3 signals was higher than centromere signals. This method also detects possible aneuploidy (polyploid cells with 3 or more centromere signals). A sample was considered NAV3 aberrant if the percentage of nuclei showing amplification/deletion exceeded the 7.8%/6.1% cut-off levels (mean ± 2 x standard deviation (2SD)) determined from the benign naevus samples, respectively.
Statistical analysis
Statistical analyses of the FISH results were performed using negative binomial regression in SPSS. The response variable in the first model was the number of nuclei with deletion, and in the second model the number of nuclei with amplification. The independent variable was the group variable, and the offset variable was the total number of counted nuclei. Multiple comparison of groups was done with Sequential Sidak adjustment method. Kaplan–Meier plot was used when evaluating the survival of the melanoma patients. Patients were divided into groups with relatively more NAV3 deletions, NAV3 amplifications, or with no change.
Statistical analyses for migration and 3D growth assays were performed with a commercially available statistical software package, IBM SPSS for Mac (IBM, Armonk, NY, USA). For qPCR, the results of NAV3 correlation with MMP14 and MMP16, Kolmogorov-Smirnov and Shapiro-Wilk tests suggested that both X and Y were non-normal. Therefore, data were log-transformed.
Quantification of mRNA expression for NAV3, MMP14 and MMP16
RNA was isolated from 10-µm tissue sections using High Pure FFPE RNA Micro Kit (Roche, Basel, Switzerland) and from WM239 cells using RNeasy Micro Kit (Qiagen, Hilden, Germany). cDNA was synthesized using SuperScript® VILO™ cDNA synthesis kit (Thermo Fisher Scientific). The TaqMan Gene Expression Assay’s probe was 20-fold and 0.5 µl was used in 1 reaction. The LightCycler performed 40 runs in each experiment. NAV3, MMP14, and MMP16 genes were amplified with iQ Supermix (control 730001045/730003323, cat. 170-8862, (Bio-Rad, Hercules, CA, USA), NAV3, Hs00372108_m1; MMP14, Hs 01037006_gH; MMP16, Hs 00234676_m1, GAPDH, Hs02786624g1, Thermo Fisher Scientific).
Sequence analysis
DNA was isolated from 10-µm tissue sections of 6 paraffin-embedded tumour samples (QIAamp DNA FFPE Tissue Kit, Qiagen) and NAV3 was amplified using gene-specific primers (forward primer 5’-ACTGCCACCAGCTCCTTTGGC-3’ and reverse primer 5’-TCTCTCAGTTTCGAGGTGGTG-3’). PCR products were purified (PCR Purification Kit, Qiagen) and sequenced at Biomedicum Sequencing Unit (Helsinki, Finland). The results were analysed using Gene Composer software (Biomedicum Bioinformatics Unit, Helsinki, Finland).
2-dimensional (2D) wound migration assay
Removable, polystyrene medium chambers of 4-well configuration Lab-Tek™ II – Chamber Slide™ System (Thermo Fisher Scientific) was used for incubation and growth of WM-239 cell line cultures. When the cell monolayer displayed approximately over 90% confluence, the monolayer was wounded with a sterile rubber blade. The wound closure was assessed 9–24 h later under light microscopy. Cells were then fixed with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for 10 min, washed with PBS, EtOH 35% ethanol (EtOH) in PBS, 50%/PBS and 70%/PBS, and immersed in 70% EtOH/PBS for storage. The distance migrated by cells was measured using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
RNA interference
Small interfering RNAs (siRNA) targeting NAV3 ((FlexiTube GeneSolution for NAV3 GS89795, siNAV3-1 (SI04141956), and siNAV3-2 (SI04272646), Qiagen) and non-silencing control siRNA (SI03650325, Qiagen) were transfected using Lipofectamine™ 2000 (Thermo Fisher Scientific). The silencing efficacies were assessed by quantitative polymerase chain reaction (qPCR).
Growth in 3D collagen
Type I collagen (4.8 mg/ml, rat tail, Sigma-Aldrich, St Louis, MO, USA) was mixed with an equal amount of 2 x minimal essential media (MEM), and the pH was adjusted to 7.4 using 20% sodium hydroxide (NaOH). A total of 5,000 cells were suspended in 40 µl hydrogel, the suspension was transferred to a 24-well plate, and incubated for 1 h at 37°C to allow complete gelling. After 48 h incubation in complete growth medium, the cultures were photographed and the percentage of elongated cells amongst the total was calculated using ImageJ software.
Immunostaining
Immunohistochemistry for NAV3 and MMP14 proteins was performed on 40 formalin-fixed and paraffin-embedded primary melanoma samples and 3 metastases, as described previously (26). After standard deparaffination, the sections were blocked with ready-to-use 2.5% normal horse blocking serum (Universal Impress, MP-7500, Vector Laboratories, Newark, CA, USA), incubated overnight at 4°C with either rabbit anti-Nav3 (HPA032111, Lot. R32215, Sigma Prestige Antibodies, Sigma-Aldrich) diluted 1:100 in 1% BSA, or mouse anti-MMP14 (MAB3328, Lot. 2450182, Millipore, Burlington, MA, USA), diluted 1:100 in 1% BSA, and further incubated with the secondary antibody Universal ImPress HRP (Universal Impress, MP-7500, Vector Laboratories) and VECTOR NovaRED (SK-4800, Vector Laboratories) substrate. The slides were counterstained with Meyer’s haematoxylin and mounted, after graded alcohol series, with Neo-Mount (HX934618, Merck, Rahway, NJ, USA). For the scratch wound slides, immunostaining was performed with the primary antibody anti-Nav3 (HPA032111, Lot. R32215, Sigma Prestige Antibodies, Sigma-Aldrich), diluted 1:300 in BSA 1%, and mouse anti-MMP14 (MAB3328, Lot. 2450182, Millipore) diluted 1:100 in BSA 1%. The slides were incubated overnight at 4°C, followed by the Vectastain imPRESS™ UNIVERSAL REAGENT anti-Mouse/Rabbit Ig peroxidase kit, (Vector Laboratories MP-7500) with Vector AEC Peroxidase Substrate Kit as the chromogen (Vector Laboratories SK-4200) and Mayer’s haematoxylin as counterstaining. Slides were mounted with Aquatex (VWR International Ltd, Lutterworth, UK). The immunostainings were analysed independently by 2 authors (OB and AR).
Double immunofluorescence staining with NAV3 and MMP14
For the double immunofluorescence, 7 thick (Breslow thickness > 2.4 mm) formalin-fixed and paraffin-embedded melanoma samples were stained for MMP14 and NAV3. Anti-MMP14 antibody (MAB3328, Lot. 2450182, Millipore) was used at 1:50 dilution in Tris-buffered saline (TBS). The secondary antibody was goat anti-mouse Alexa 594, diluted 1:500 in TBS. The second primary antibody was the same anti-NAV3 antibody as above, used at a dilution of 1:100 in TBS. The primary antibody was incubated overnight at 4°C, and goat anti-rabbit Alexa 488 was used as the secondary antibody, diluted 1:500 in TBS. To visualize the cell nuclei (DNA), the sections were treated with Hoechst 33342 solution (Thermo Fisher Scientific) and the slides were mounted with Immu-mount (Thermo Fisher Scientific). The images were visualized using immunofluorescence Zeiss Axio Imager microscope (Carl Zeiss Ag, Oberkochen, Germany).
RESULTS
NAV3 aberrations were found in most primary melanomas but not in benign naevi
To determine the possible role of NAV3 in melanoma progression, this study analysed NAV3 (chromosome 12q21) copy number alterations with the use of FISH in primary melanomas and benign naevi. In the primary melanomas, NAV3 copy number changes were observed in 18/27 (67%) tumours, while no copy number alterations were found in benign naevi (Table I). Chromosome 12 polysomy was found in 13/27 (48%) of primary melanomas but not in benign nevi. NAV3 deletions were found in 16/27 (59%) of the primary melanomas, and, in 5 of these, the proportion of nuclei with deletions was very high, ranging from 46% to 97% (Table I). NAV3 amplification was found in 12/27 (44%) of the primary melanomas, accompanied by chromosome 12 polysomy in 5 cases. NAV3 amplification was seen in 8–20% of the tumour cells at most, and 10 tumours (37%) showed heterogeneity in NAV3 copy number so that both types of aberrated nuclei were present. The proportion of NAV3 deletions in primary melanomas showed statistically significant difference compared with benign naevi (p = 0.02; Table I). The differences in frequency of NAV3 amplifications did not reach statistical significance.
| Sample and diagnosis | NAV3 deletiona | NAV3 amplificationa | Chromosome 12 polysomy |
| Melanoma (n = 27) | 16/27 (59%)* | 12/27 (44%) | 13/27 (48%) |
| Benign naevus (n = 15) | 0/15 (0%) | 0/15 (0%) | 0/15 (0%) |
| WM-793 primary melanoma cell line | 36 ± 6% | 20 ± 9% | 83 ± 7% |
| WM-239 metastatic melanoma cell line | 21 ± 7% | 9 ± 6% | 97 ± 1% |
| aA sample was considered NAV3 aberrant if the percentage of nuclei showing amplification/deletion exceeded the mean +2 SD determined from the benign naevus samples. The cut-off was 6.1% for deletion and 7.8% for amplification. *p = 0.02. | |||
The 2 melanoma cell lines were both heterogenic, since 20% of cells in the primary WM-793 cell culture showed amplifications and 36% of cells showed deletions of NAV3, whereas in the metastasis-derived WM-239 cell culture 9% of cells showed amplifications and 21% of cells showed deletions (Table I).
Since NAV3 has previously been shown to be mutated in melanoma samples with a nucleotide change c.598C > T (4), the current study analysed the corresponding NAV3 mutations in the melanoma patient samples (6 patients), but could not detect this transition in any of the samples (data not shown).
NAV3 protein expression localizes to the leading edge of migrating melanoma cells in vitro
To understand the role of NAV3 in melanoma cell migration, a 24-h 2-dimensional (2D) scratch wound assay was generated using the WM-239 melanoma cell line. The assay revealed different intracellular localization and patterns of NAV3 staining in the melanoma cells according to the location of the cells, either on the border of the wound or distant from the wound (Fig. 1). A striking unilateral polarization of NAV3 was seen at the leading edge of migrating cells (Fig. 1A, B), while no polarization of NAV3 staining was seen in the non-scratched area (Fig. 1C, D). Migrating melanoma cells disclosed the morphology of migrating malignant cells, such as bi- or tri-polar dendritic melanocytes (27).

Fig. 1. NAV3 is polarized to the direction of cell movement in WM-239 melanoma cells. (A, B) NAV3 immunohistochemistry (red) of a WM-239 cell culture at the border of a scratched area after 24 h. A unilateral polarization of NAV3 is seen to the direction of cell movement (arrows). (C, D) NAV3 immunohistochemistry of a non-scratched area. NAV3 staining at the cellular membrane is not polarized (arrowheads). (D) Enlargement of the area marked in (C). Scale bar: 100 µm (A, C); 50 µm (B and D).
Silencing of NAV3 reduces melanoma cell migration in 2D environment and invasive growth in 3D type I collagen
To assess the role of NAV3 in melanoma cell motility, the current study silenced NAV3 from WM239 melanoma cells using specific siRNAs (Fig. 2A). In a 9-h 2D wound scratch migration assay, NAV3 silencing with the most efficient siRNA, siNAV3-2, reduced the distance migrated by cells by over 50% (Fig. 2B). While control cells exhibited elongated morphology at the migration front, cells silenced for NAV3 remained rounded (Fig. 2C), suggesting that NAV3 is also necessary for microtubule stabilization in melanoma cells.

Fig. 2. Silencing NAV3 reduces melanoma cell migration and invasive phenotype in collagen. (A) Relative NAV3 mRNA expression in WM-239 cells transfected with control siRNA (siCtrl) and siRNA targeting NAV3 (siNAV3-1 and siNAV3-2) as assessed by qPCR. (B) Relative distance migrated by WM-239 cells transfected with siCtrl and siNAV3-1 and siNAV3-2 in a 9-h wound scratch migration assay. (C) Light micrographs visualize WM-239 cells transfected with siCtrl and siNAV3-2 at the time points 0 h and 9 h in wound scratch migration assay. Scale bar 200 µm (D) WM-239 cells transfected with siCtrl and siNAV3-1 and siNAV3-2 were grown in 3-dimensional (3D) type I collagen for 48 h. Chart shows the percentage of elongated cells among the total, which indicates the invasive ability of the cells. (E) Light micrographs visualize WM-239 cells grown in 3D collagen type I for 48 h. Scale bar 400 µm *p < 0.05, **p < 0.001.
When the cells were embedded into dense 3D type I collagen, 50% of WM239 cells grown in 3D collagen showed elongated cell morphology and sprouting, reflective of invasive activity (Fig. 2D and E). Notably, silencing of NAV3 markedly reduced the cell elongation and invasive phenotype (Fig. 2D and E), suggesting that NAV3 is required for invasive melanoma cell invasive capability inside dense collagen type I.
NAV3 expression in primary melanoma tumours
To further investigate NAV3 functions in melanoma, the current study assessed NAV3 protein expression in human melanoma tumours using immunohistochemistry in 39 primary melanomas. NAV3 expression was observed in 27/39 of the melanoma tumours (Fig. 3). Notably, NAV3 was strongly and homogenously expressed in all cells of every sample of thin tumours (8/8, Breslow thickness < 1 mm, Fig. 3). In thicker tumours (1–7 mm), in turn, NAV3 was expressed in 19/31 (61%) tumours. In the mid-thickness samples (1–5 mm), NAV3 was expressed in 16/24 (67%) tumour samples. The expression pattern in mid-thickness tumours was heterogeneous, with only a proportion of the tumour cells expressing NAV3, either at the epidermal edge or at the invasive edge of the tumours (Fig. 3). In 7/7 of the thickest tumours (> 5 mm) NAV3 was not expressed (4/7) or was expressed only weakly in the upper dermis, but lost from the lower parts of the dermis (3/7, Fig. 3).
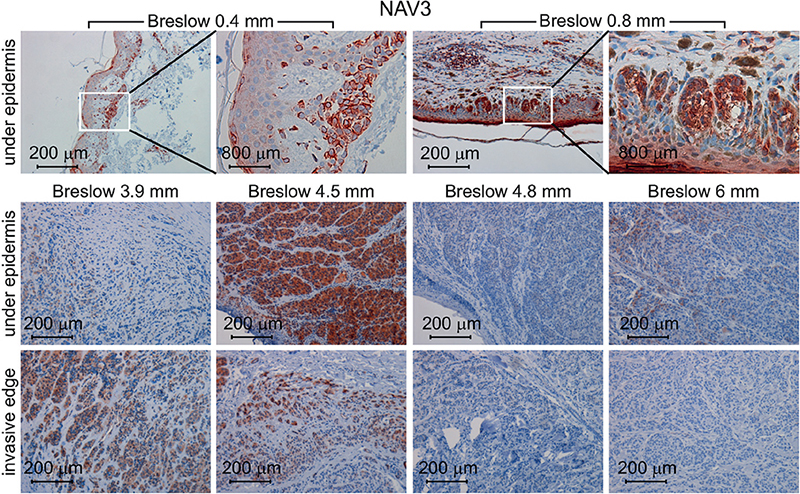
Fig. 3. NAV3 expression is downregulated with increase in melanoma thickness. NAV3 immunohistochemistry of human primary melanoma samples visualizes its expression. NAV3 was expressed in all cells of thin melanomas (Breslow thickness < 1 mm), while in thicker melanomas it was expressed either under the epidermis or at the invasive edge, while in the thickest melanomas (Breslow thickness > 5 mm) it was often lost.
NAV3 and MMP14 co-expression in primary melanoma tumours
Since NAV3 was polarized to the leading edge of migrating melanoma cells and its silencing reduced invasive growth inside 3D collagen, the current study explored the possible co-expression of NAV3 with the membrane-tethered proteinase MMP14, which is major type I collagenase, which is required for invasive growth in 3D collagen and is also expressed at the leading edge of invading melanoma cells. In addition, this study assessed the expression of the close homologue of MMP14, MMP16, which modulates MMP14 function and is associated with poor prognosis in melanoma (19, 22, 26). mRNA expression of these proteins was analysed in 13 primary tumours, and expression of NAV3 correlated positively with MMP14 (r = 0.85, p = 0.0022), but not with MMP16 (r = 0.35, p = 0.041, Fig. 4A and 4B).

Fig. 4. NAV3 mRNA expression correlates with MMP14 mRNA in melanoma. Relative mRNA expression of NAV3 and MMP14 (A, p = 0.00218) and MMP16 (B, p = 0.04114) in 13 primary melanomas was assessed by qPCR. r = Pearson’s correlation.
To further elucidate the association of NAV3 and MMP14 proteins, the current study tested for NAV3 and MMP14 co-expression, using immunohistochemistry and immunofluorescence staining of primary melanoma tumours. NAV3 and MMP14 were similarly expressed (i.e. either both proteins were expressed or were not expressed in the same sample) in 26/37 (70%) of all samples. Both proteins were expressed in all thin melanoma tumours with Breslow thickness < 1 mm (8/8, 100%), while of the mid-thickness tumours (1–5 mm) 11/23 (48%), and of the thickest tumours (5–7 mm) only 1/6 were positive for both NAV3 and MMP14 (Table II, Fig. 5). The 12 tumours that were negative for either NAV3 alone (n = 6) or for both NAV3 and MMP14 (n = 6) were all > 1 mm. Notably, 4/6 of the double-negative tumours were ≥ 6 mm thick. NAV3 and MMP14 co-localized to the same cells in all double-positive samples (Fig. 5A and B). Consistently, NAV3 deletion or downregulation (16, 17), as well as MMP14 downregulation (26, 28) have been reported in metastatic tumours of many cancer types. Collectively, these results show that NAV3 is often downregulated together with MMP14 in thick melanoma tumours, suggesting that NAV3 and MMP14 downregulation may favour melanoma tumour growth and/or dissemination.

Fig. 5. NAV3 and MMP14 are co-expressed in the same cells in primary melanoma tumours. (A) Fluorescence micrographs visualize NAV3/MMP14 co-expression in the melanoma tumours (upper and middle rows). Four out of 6 very thick ≥ 6 mm tumours were double negative for NAV3 and MMP14 (bottom row). (B) NAV3 and MMP14 were expressed only at the invasive edge of some thick tumours. NAV3 (green), MMP14 (red), DAPI (blue).
DISCUSSION
Malignant transformation is thought to arise due to a series of chromosomal aberrations and somatic mutations, which finally lead to a cell phenotype that has the capacity for uncontrolled proliferation and metastasis. In the spread of melanoma, both the motility of cells and their ability to modulate extracellular matrix are required for cancer cell invasion into deeper dermis and blood and lymphatic vessels (26, 29). Cancer cells use different types of invasion dependent on the microenvironment: protease-independent amoeboid invasion, in which cells squeeze through pre-existing spaces between fibres; or mesenchymal invasion, in which cells have invadopodia and protrude by degrading extracellular matrix (29). NAV3, a microtubule plus-end tracking protein and tumour suppressor, regulates cell migration and invasion by stabilizing microtubules needed for invadopodia formation, and inhibits metastasis in breast cancer and colon cancer (3, 16, 17). Membrane-type matrix metalloproteinase MMP14 is a major collagenase that confers cancer cells with an ability to degrade extracellular matrix and invade in a mesenchymal manner (30). The current study shows that, in human melanoma, NAV3 promotes directional melanoma cell migration, elongated morphology, and invasive growth in 3D collagen, and is strongly expressed in all early-stage melanomas together with MMP14. However, NAV3 and MMP14 protein expression is often downregulated or lost with tumour progression from thin to mid-thick and, further, to very thick melanomas, suggesting that their loss favours tumour spreading in melanoma.
NAV3 deletion is associated with poorer outcome in cancer patients, and NAV3 silencing enhances metastases in breast cancer cells, probably by reducing apoptosis and reducing directional migration of tumour cells (16). Notably, NAV3 was recently shown to be a downstream target of SOX9 (31), which, in turn, has been shown to induce melanoma cell invasion and metastasis (32). The current study found that 59% of primary melanomas had NAV3 deletion. Moreover, NAV3 silencing in melanoma cells reduced their migration in a wound scratch assay and changed the morphology of migrating cells from elongated to rounded, as has also been seen in breast cancer cells (16). In addition, NAV3 was expressed at the tips of invadopodia of migrating melanoma cells, and its silencing reduced sprouting of melanoma cells in the 3D collagen, suggesting that NAV3 promotes melanoma cell motility both in the 2D and the 3D environment.
Since MMP14 is a major mediator of protease-driven invasion inside type I collagen (30) and is upregulated in melanoma (33), it was decided to explore the correlation of its expression with NAV3. Blocking the protease activity of MMP14 with a monoclonal antibody in a melanoma mouse xenograft model alleviated tumour metastatic burden (34), and MMP-based interventions are under investigation for cancer treatment. However, the function of MMP14 is more complex than being merely a tumour promoter. Previously we have shown that limiting the functional MMP14 protein in melanoma cell surfaces leads to a more aggressive phenotype and lymphatic invasion by switching the melanoma invasion pattern from infiltrating to expansive, whereby induced cell-cell contacts and reduced collagen degradation by tumour cells leads to expansive growth of melanoma cell nests and lymphatic invasion of the collective cell clusters (26). Cepeda et al. also report that lower MMP14 expression is associated with increased migratory capacity and tumourigenicity in a breast cancer model compared with high MMP14-expressing cells (28).
The current study found that NAV3 mRNA expression correlated with MMP14 mRNA in human melanoma samples. NAV3 and MMP14 proteins were either co-expressed or their expression was lost in 26/37 (70%) of melanoma samples, suggesting the common regulation of these proteins. In addition, this study found that, at the early stage of melanoma, NAV3 and MMP14 were strongly expressed in all tumours (thin tumours with Breslow thickness under 1 mm). Of note, both NAV3 and MMP14 became weaker or completely undetectable in thicker tumours, which represent later stages of melanoma progression, suggesting that NAV3 and MMP14, although important at the beginning of tumour growth and invasion in collagen-rich and dense upper dermis, are not necessary or can even restrict tumour growth after the tumour has reached a certain size or permissive immediate microenvironment.
Although the potential co-regulation mechanisms and functional collaboration of MMP14 and NAV3 proteins remain to be explored, there are several mechanisms that may explain the similar expression pattern of these 2 proteins. Being a plus-end protein of the microtubules, NAV3 may affect MMP14 localization to the invadopodia, since microtubules are required for MMP14 trafficking to these structures (23). Furthermore, NAV3 regulates epidermal growth factor receptor (EGFR) endocytosis, and epidermal growth factor (EGF) is an important inducer of MMP14 expression in ovarian cancer cells at least (35). MMP14, in turn, promotes the release of heparin-binding EGF-like growth factor (HB-EGF) (36), which induces NAV3. Taken together, these results show that NAV3 deletion is a common feature in human melanoma, and that, although NAV3 is required for the invasive growth of early melanoma and is strongly expressed at the early stages of melanoma, it is lost in most melanoma tumours at later stages, suggesting that NAV3 may serve as a marker of melanoma progression.
ACKNOWLEDGEMENTS
The authors thank Kaija Järvinen, Alli Tallqvist and Anastasia Chernenko for excellent technical assistance, and the Biomedicum Imaging Unit (Biomedicum Helsinki) for providing imaging facilities.
REFERENCES
- Rager EL, Bridgeford EP, Ollila DW. Cutaneous melanoma: update on prevention, screening, diagnosis, and treatment. Am Fam Physician 2005; 72: 269–276.
- Leonardi G, Candido S, Falzone L, Spandidos DA, Libra M. Cutaneous melanoma and the immunotherapy revolution (Review). Int J Oncol 2020; 57: 609–618.
- van Haren J, Draegestein K, Keijzer N, Abrahams JP, Grosveld F, Peeters PJ, et al. Mammalian Navigators are microtubule plus-end tracking proteins that can reorganize the cytoskeleton to induce neurite-like extensions. Cell Motil Cytoskeleton 2009; 66: 824–838.
- Bleeker FE, Lamba S, Rodolfo M, Scarpa A, Leenstra S, Vandertop WP, et al. Mutational profiling of cancer candidate genes in glioblastoma, melanoma and pancreatic carcinoma reveals a snapshot of their genomic landscapes. Hum Mutat 2009; 30: E451–459.
- Carlsson E, Ranki A, Sipilä L, Karenko L, Abdel-Rahman WM, Ovaska K et al. Potential role of a navigator gene NAV3 in colorectal cancer. Br J Cancer 2012; 106: 517–524.
- Coy JF, Wiemann S, Bechmann I, Bächner D, Nitsch R, Kretz O, et al. Pore membrane and/or filament interacting like protein 1 (POMFIL1) is predominantly expressed in the nervous system and encodes different protein isoforms. Gene 2002; 290: 73–94.
- Hahtola S, Burghart E, Jeskanen L, Karenko L, Abdel-Rahman WM, Polzer B, et al. Clinicopathological characterization and genomic aberrations in subcutaneous panniculitis-like T-cell lymphoma. J Invest Dermatol 2008a; 128: 2304–2309.
- Hahtola S, Burghart E, Puputti M, Karenko L, Abdel-Rahman WM, Väkevä L, et al. Cutaneous T-cell lymphoma-associated lung cancers show chromosomal aberrations differing from primary lung cancer. Genes Chromosomes Cancer 2008b; 47: 107–117.
- Karenko L, Hahtola S, Päivinen S, Karhu R, Syrjä S, Kähkönen M, et al. Primary Cutaneous T-Cell Lymphomas Show a Deletion or Translocation Affecting NAV3, the Human UNC-53 Homologue. Cancer Res 2005; 65: 8101–8110.
- Nord H, Hartmann C, Andersson R, Menzel U, Pfeifer S, Piotrowski A, et al. Characterization of novel and complex genomic aberrations in glioblastoma using a 32K BAC array. Neuro Oncol 2009; 11: 803–818.
- Soon PSH, Gill AJ, Benn DE, Clarkson A, Robinson BG, McDonald KL, et al. Microarray gene expression and immunohistochemistry analyses of adrenocortical tumors identify IGF2 and Ki-67 as useful in differentiating carcinomas from adenomas. Endocr Relat Cancer 2009; 16: 573–583.
- Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science 2007; 318: 1108–1013.
- Zhang L, Luo M, Yang H, Zhu S, Cheng X, Qing C. Next-generation sequencing-based genomic profiling analysis reveals novel mutations for clinical diagnosis in Chinese primary epithelial ovarian cancer patients. J Ovarian Res 2019; 12: 19.
- Aly J, Lewis T, Parikh T, Britten J, Malik M, Catherino W. NAV3, a tumor suppressor gene, is decreased in uterine leiomyoma tissue and cells. Reprod Sci 2020; 27: 925–934.
- Carlsson E, Krohn K, Ovaska K, Lindberg P, Häyry V, Maliniemi P et al. Neuron navigator 3 alterations in nervous system tumors associate with tumor malignancy grade and prognosis. Genes Chromosomes Cancer 2013; 52: 191–201.
- Cohen-Dvashi H, Ben-Chetrit N, Russell R, Carvalho S, Lauriola M, Nisani S, et al. Navigator-3, a modulator of cell migration, may act as a suppressor of breast cancer progression. EMBO Mol Med 2015; 7: 299–314.
- Uboveja A, Satija Y, Siraj F, Sharma I, Saluja D. p73 – NAV3 axis plays a critical role in suppression of colon cancer metastasis. Oncogenesis 2020; 9: 12.
- Napoli S, Scuderi C, Gattuso G, Di Bella V, Candido S, Basile MS, et al. Functional roles of matrix metalloproteinases and their inhibitors in melanoma. Cells 2020; 9: 1151.
- Szabova L, Chrysovergis K, Yamada SS, Holmbeck K. MT1-MMP is required for efficient tumor dissemination in experimental metastatic disease. Oncogene 2008; 27: 3274–3281.
- Hofmann UB, Houben R, Bröcker E-B, Becker JC. Role of matrix metalloproteinases in melanoma cell invasion. Biochimie 2005; 87: 307–314.
- Raeeszadeh-Sarmazdeh M, Do LD, Hritz BG. Metalloproteinases and their inhibitors: potential for the development of new therapeutics. Cells 2020; 9: 1313.
- Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, et al. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol 2003; 160: 267–277.
- Castro-Castro A, Janke C, Montagnac G, Paul-Gilloteaux P, Chavrier P. ATAT1/MEC-17 acetyltransferase and HDAC6 deacetylase control a balance of acetylation of alpha-tubulin and cortactin and regulate MT1-MMP trafficking and breast tumor cell invasion. Eur J Cell Biol 2012; 91: 950–960.
- Maliniemi P, Carlsson E, Kaukola A, Ovaska K, Niiranen K, Saksela O et al. NAV3 copy number changes and target genes in basal and squamous cell cancers. Exp Dermatol 2011; 20: 926–931.
- Ranki A, Väkevä L, Sipilä L, Krohn K. Molecular markers associated with clinical response to bexarotene therapy in cutaneous T-cell lymphoma. Acta Derm Venereol 2011; 91: 568–573.
- Tatti O, Gucciardo E, Pekkonen P, Holopainen T, Louhimo R, Repo P, et al. MMP16 mediates a proteolytic switch to promote cell-cell adhesion, collagen alignment, and lymphatic invasion in melanoma. Cancer Res 2015; 75: 2083–2094.
- Cobb JP, Walker DG. Studies on human melanoma cells in tissue culture. I. Growth characteristics and cytology. Cancer Res 1960; 20: 858–867.
- Cepeda MA, Pelling JJ, Evered CL, Williams KC, Freedman Z, Stan I, et al. Less is more: low expression of MT1-MMP is optimal to promote migration and tumourigenesis of breast cancer cells. Mol Cancer 2016; 15: 65.
- Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr Opin Cell Bio 2004; 16: 14–23.
- Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol 2004; 167: 769–781.
- Raza S, Jokl E, Pritchett J, Martin K, Su K, Simpson K. SOX9 is required for kidney fibrosis and activates NAV3 to drive renal myofibroblast function. Sci Signal 2021; 14: eabb4282.
- Cheng PF, Shakhova O, Widmer DS, Eichhoff OM, Zingg D, Frommel SC, et al. Methylation-dependent SOX9 expression mediates invasion in human melanoma cells and is a negative prognostic factor in advanced melanoma. Genome Biol 2015; 16: 42.
- Hofmann U, Westphal J, Van Muijen G, Ruiter D. Matrix metalloproteinases in human melanoma. J Invest Dermatol 2000; 115: 337–344.
- Remacle A, Cieplak P, Nam D, Shiryaev S, Ge X, Strongin A. Selective function-blocking monoclonal human antibody highlights the important role of membrane type-1 matrix metalloproteinase (MT1-MMP) in metastasis. Oncotarget 2017; 8: 2781–2799.
- Cowden Dahl KD, Zeineldin R, Hudson LG. PEA3 is necessary for optimal epidermal growth factor receptor-stimulated matrix metalloproteinase expression and invasion of ovarian tumor cells. Mol Cancer Res 2007; 5: 413–421.
- Overland A, Insel P. Heterotrimeric G proteins directly regulate MMP14/membrane type-1 matrix metalloprotease: a novel mechanism for GPCR-EGFR transactivation. J Biol Chem 2015; 290: 9941–9947.