ORIGINAL REPORT
Antimicrobial Peptide Loss, Except for LL-37, is not Characteristic of Atopic Dermatitis
Lilla SZABÓ1,2†, Anikó KAPITÁNY1,3†, Orsolya SOMOGYI1–3, Iman ALHAFEZ1, Krisztián GÁSPÁR1, Réka PALATKA1,2, Lilla SOLTÉSZ1,2, Dániel TÖRŐCSIK1,3, Zoltán HENDRIK4, Zsolt DAJNOKI1# and Andrea SZEGEDI1,3#
1Department of Dermatology, Centre of Excellence, Faculty of Medicine, University of Debrecen, Debrecen, Hungary, 2Gyula Petrányi Doctoral School of Allergy and Clinical Immunology, University of Debrecen, Debrecen, Hungary, 3ELKH-DE Allergology Research Group, Debrecen, Hungary and 4Department of Forensic Medicine, Faculty of Medicine, University of Debrecen, Debrecen, Hungary
†These authors contributed equally to this work. #These authors contributed equally to this work.
Atopic dermatitis is an inflammatory skin disease characterized by significant permeability barrier damage. Regulation and maintenance of permeability and antimicrobial skin barriers are strongly connected. There is a lack of comprehensive studies of the expression of all 5 major antimicrobial peptide functional groups in atopic dermatitis. The aim of this study was to investigate the major antimicrobial peptide functional groups in lesional atopic dermatitis, non-lesional atopic dermatitis, and healthy control samples, using real-time quantitative PCR and immunohistochemistry. Lesional psoriatic skin was also examined as a diseased control. No differences in mRNA levels were detected between non-lesional atopic dermatitis and healthy control skin, and, at the protein level, the only change was the significantly decreased LL-37 in non-lesional atopic dermatitis. In lesional atopic dermatitis, several antimicrobial peptides were significantly altered at the mRNA level, while, at the protein level, all antimicrobial peptides were significantly upregulated or unchanged, except for LL-37, which decreased, compared with healthy controls. Antimicrobial peptides were similarly elevated in lesional atopic dermatitis and lesional psoriatic skin, with somewhat higher expression in lesional psoriatic skin, except for LL-37. In conclusion, LL-37 was the only antimicrobial peptide that was impaired in both non-lesional and lesional atopic dermatitis, highlighting its potential pathogenetic or exacerbating role in the initial stages of the disease.
Key words: antimicrobial peptide; atopic dermatitis; skin barrier; psoriasis.
SIGNIFICANCE
Data regarding antimicrobial peptides in atopic dermatitis are incomplete, and many discrepancies exist, which may be because the expression of antimicrobial peptides has often been compared with psoriatic rather than control skin. This study comprehensively analysed the main antimicrobial peptide representatives, at both the mRNA and protein levels, in clinically asymptomatic and symptomatic skin from patients with atopic dermatitis, and examined diseased control (psoriatic) and healthy control samples. The facts that the only impairment was a lack of induction of LL-37, and that LL-37 is associated with all the major pathogenic features of atopic dermatitis, indicate a driver role for LL-37 in the pathophysiology of atopic dermatitis, and raise the possibility of its therapeutic potential.
Citation: Acta Derm Venereol 2023; 103: adv9413. DOI https://doi.org/10.2340/actadv.v103.9413.
Copyright: © Published by Medical Journals Sweden, on behalf of the Society for Publication of Acta Dermato-Venereologica. This is an Open Access article distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (https://creativecommons.org/licenses/by-nc/4.0/)
Accepted: May 30, 2023; Published: Jun 30, 2023
Corr: Andrea Szegedi, Department of Dermatology, Faculty of Medicine, University of Debrecen, HU-4032 Debrecen, Hungary. E-mail: aszegedi@med.unideb.hu
Competing interests and funding: The authors have no conflicts of interest to declare.
Hungarian Research Grants (NKFIH K-128250, NKFIH K-142348, and NKFIH PD-131689), EFOP-3.6.1-16-2016-00022 project, János Bolyai Research Scholarship of the Hungarian Academy of Sciences, and ÚNKP-22-3 New National Excellence Program of the Ministry for Culture and Innovation
INTRODUCTION
The skin is the primary line of defence of the human body. In addition to providing a physical barrier, the skin senses, transmits, and responds to signals from the outside world via its antimicrobial and immunological barriers (1, 2). The antimicrobial barrier is formed by antimicrobial peptides (AMPs), which have both antimicrobial activity and immunomodulatory effects under homeostatic and inflammatory conditions (3). AMPs are classified into 5 groups based on their functions: classic AMPs, AMPs with protease inhibitor or enzymatic activity, AMPs with chemokine activity, AMPs with neuropeptide activity, and AMPs that do not fit into any other group (4).
AMPs are key factors in the pathogenesis of several immune-mediated skin diseases, including Th1/Th17-driven psoriasis vulgaris (PsV) and rosacea (5). AMP levels are notably increased in these skin diseases. In contrast, the role of AMPs in the pathogenesis of Th2/Th22-driven atopic dermatitis (AD) is less obvious, there are several uncertainties in this topic. Some contradictions have arisen because AD has been compared with PsV samples without healthy controls in several studies. Furthermore, only mRNA levels were measured in some studies, non-lesional AD (AD NL) samples were not involved in many studies, and the role of AMP functional groups in AD was not examined.
The aim of the current study was to comprehensively analyse AMPs in AD NL and lesional AD (AD L) skin compared with healthy controls. Lesional PsV (PsV L) samples were also analysed as diseased controls. Expression of the keratinocyte-expressed representative members of all 5 main functional AMP groups was investigated at the mRNA level by real-time quantitative PCR (RT-qPCR) and at the protein level by immunohistochemistry (IHC) combined with quantification following whole-slide imaging.
MATERIALS AND METHODS
Skin biopsies
Biopsies were collected from the lesional and non-lesional skin of 10 AD patients with chronic symptoms, from the lesional skin of PsV patients, and from the corresponding skin regions of 10 healthy individuals (Table I), as distinct healthy skin regions have different immune activity (2, 6, 7). Written, informed consent was obtained, according to the principles of the Declaration of Helsinki, and the study was approved by the local ethics committee (Regional Institutional Research Ethics Committee, Clinical Center, University of Debrecen, Debrecen, Hungary; study i.d.: IV/2072-2/2020/EKU). One part of the biopsies was stored in RNAlater (Qiagen, Hilden, Germany) at –70°C until RNA isolation for RT-qPCR, the other part of the biopsies was formalin-fixed and paraffin-embedded and used for IHC.
Real-time quantitative PCR
Sample preparation and reactions were performed as described previously (2, 6). The oligo sets used are shown in Appendix S1.
Immunohistochemistry
Freshly prepared paraffin-embedded sections of skin from AD and PsV patients and healthy controls were used. IHC experiments and quantification were performed as described previously (2, 6). The primary and secondary antibodies used are shown in Appendix S1.
Statistical analysis
Statistical significance was determined by 1-way analysis of variance (ANOVA) and Newman-Keuls post hoc tests. Graphs show the means and the corresponding 95% confidence intervals (95% CI) (boxes) and maximum/minimum values of protein levels (Figs 1 and 2).
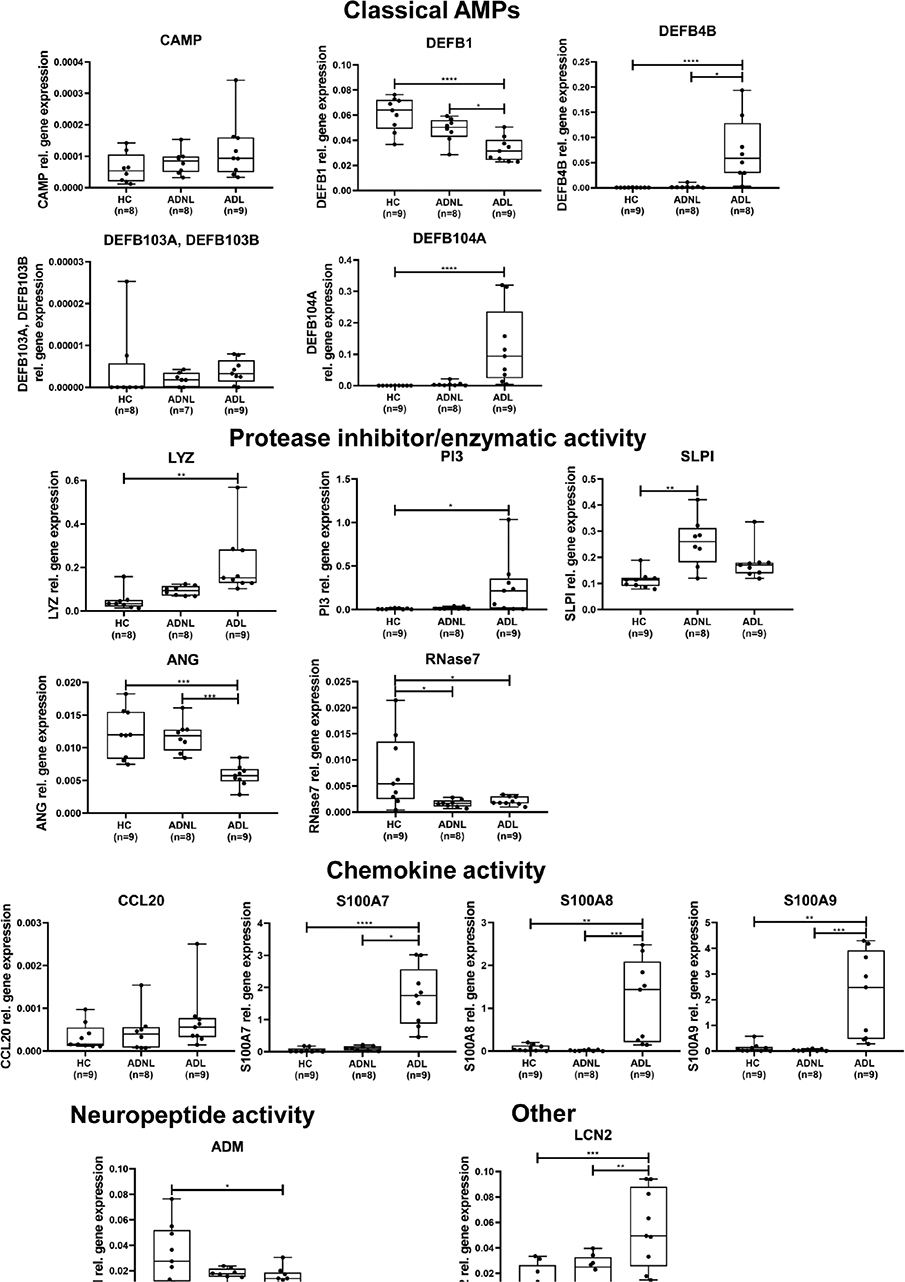
Fig. 1. Antimicrobial peptide (AMP) mRNA levels detected by real-time quantitative PCR (RT-qPCR) in atopic dermatitis lesional (AD L), AD non-lesional (NL), and healthy control (HC) skin samples AMPs are classified into functional groups. The graphs show the median ±95% confidence interval (95% CI) (*p < 0.05; **p < 0.01; ***p < 0.001, determined by 1-way analysis of variance (ANOVA) followed by Tukey’s post hoc test in case of normal distribution or Kruskal–Wallis test followed by Dunn’s post hoc test when data distribution was not normal). DEFB: defensin beta; CAMP: cathelicidin antimicrobial peptide; LYZ: lysozyme; ANG: angiogenin; SLPI: secretory leukocyte peptidase inhibitor; CCL: chemokine (C-C motif) ligand; S100: S100 calcium-binding protein; ADM: adrenomedullin; LCN. lipocalin.
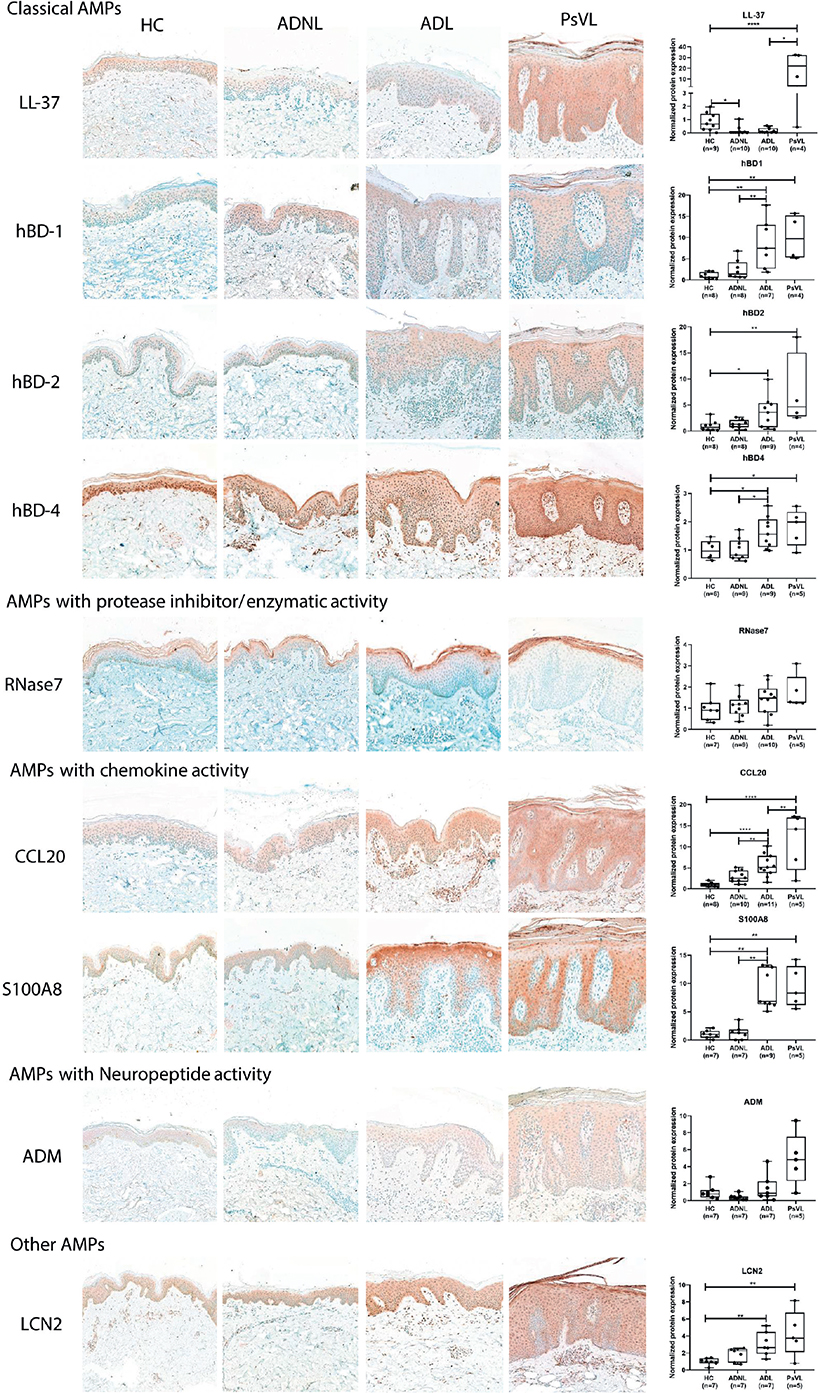
Fig. 2. Representative images for immunostaining and epidermal quantification of antimicrobial peptides (AMPs) classified into functional groups in atopic dermatitis lesional AD L, AD non-lesional (NL), psoriasis vulgaris lesional (PsV L), and healthy control (HC) skin samples. The graphs show the median ± 95% confidence interval (95% CI) of measured protein levels (*p < 0.05; **p < 0.01; ***p < 0.001, as determined by 1-way analysis of variance (ANOVA) followed by Sidak’s post hoc test in case of normal distribution or Kruskal–Wallis test followed by Dunn’s post hoc test when data distribution was not normal). hBD: human beta-defensin; CCL: chemokine (C-C motif) ligand; S100: S100 calcium-binding protein; ADM: adrenomedullin; LCN: lipocalin. original magnification: 200x.
RESULTS
Antimicrobial peptide mRNA expression in non-lesional atopic dermatitis and lesion atopic dermatitis skin
First, the study detected and compared the mRNA levels of AMPs in AD NL and healthy control skin samples. Significant change was barely detectable; only 2 AMPs were found to be significantly differentially expressed at the mRNA level, namely RNASE7 with enzymatic activity was downregulated, while secretory leukocyte peptidase inhibitor (SLPI) was expressed at higher levels in AD NL samples vs healthy controls (Fig. 1, Table SI). Other AMPs were present at similar levels in the 2 sample groups (Fig. 1, Table SI).
Next, AD L samples were compared with control skin. The classic AMPs, the expression of DEFB4B and DEFB104A (encoding human beta defensin (hBD)-2 and hBD-4, respectively) were significantly higher in AD L skin compared with control skin (Fig. 1, Table SI). In contrast, DEFB1 (encoding hBD-1) mRNA showed an opposite trend, with significant differences between AD L and control skin. Gene expression levels of DEFB103A/DEFB103B and CAMP (encoding hBD-3 and cathelicidin/LL-37, respectively) were not significantly different between the sample groups (Fig. 1, Table SI). Regarding AMPs with protease inhibitor or enzymatic activity, peptidase inhibitor 3 (PI3) and lysozyme (LYZ), were significantly higher, while angiogenin (RNASE5/ANG) and RNASE7 mRNA levels were significantly lower in AD L skin compared with the levels in healthy controls. SLPI gene expression levels were similar in AD L and healthy skin (Fig. 1, Table SI). Regarding AMPs with chemokine activity, the mRNA expression levels of S100 calcium-binding protein A molecules were significantly higher in AD L skin compared with the levels in control skin (Fig. 1, Table SI). No significant difference in C-C motif chemokine ligand (CCL)20 was observed between the sample groups. The gene expression levels of adrenomedullin (ADM), which has neuropeptide activity, were significantly lower in AD L skin vs controls. Finally, lipocalin-2 (LCN2) gene expression levels were significantly elevated in AD L skin (Fig. 1, Table SI).
Protein expression of antimicrobial peptides in non-lesional atopic dermatitis and lesion atopic dermatitis skin
As proteins are the functional forms of molecules, and mRNA and protein expression do not always coincide due to transcriptional modifications, representatives of all AMP functional groups were subsequently determined and quantified at the protein level, using IHC following whole-slide imaging.
When comparing AD NL and control groups, in line with the mRNA findings, similar protein levels were detected for almost all AMPs (hBDs, RNase7, CCL20, S100A8, ADM, LCN2), except LL-37 levels, which were significantly lower in AD NL skin compared with control skin (Fig. 2, Table SII).
When comparing AD L and healthy controls, most investigated AMPs (6 of 9) were significantly upregulated in AD L skin compared with healthy control skin, in agreement with the mRNA results (Fig. 2, Table SII). Two AMPs were present at similar levels, while only 1 AMP was significantly lower in AD L skin compared with control skin. Concerning classic AMPs, hBDs were present at significantly higher levels, while LL-37 was significantly downregulated in AD L skin compared with controls (Fig. 2, Table SII). The protein levels of CCL20 and S100A8 were significantly higher in AD L skin compared with healthy control skin. No significant differences in RNase7 and ADM protein levels were detected between the AD L and healthy skin samples. However, LCN2 level was significantly higher in AD L skin vs controls (Fig. 2, Table SII).
Regarding the staining pattern of the investigated AMPs, it was found that LCN2, hBD-4, ADM, and hBD-4 were homogeneous. LCN2 and hBD-4 staining was strong in AD L samples, while ADM staining was weak and LL-37 staining was barely detectable in AD samples. Concerning hBD-1, hBD-2, S100A8, and LCN2, they were localized mainly in the apical part of the epidermis, with decreasing levels towards the basal keratinocytes in all sample groups. Strong staining for RNase7 was observed in the stratum corneum and the upper granular epidermal layers in all sample groups.
In summary, only LL-37 showed significantly decreased levels in AD NL vs controls, while other AMPs were not induced. AMPs with significantly decreased mRNA levels did not show a decrease at the protein level in AD L samples. AMP protein levels in AD L were significantly increased in most cases, 2 were unchanged, and LL-37 was the only AMP that was lower in AD L vs control.
Comparison of antimicrobial peptide protein levels in lesion atopic dermatitis and lesional psoriasis vulgaris skin
It was hypothesized that some inconsistencies in the literature may be due to AD AMP levels being compared with PsV skin instead of healthy skin. Thus, immunostainings of AMPs in PsV L samples were performed to clarify this discrepancy.
It was found that PsV L skin had highly and significantly elevated protein expression compared with the control skin for most AMPs. ADM and RNase7 exhibited prominent staining, but the levels were not significantly different in PsV L skin compared with healthy control skin (Fig. 2).
When comparing AD L and PsV L, interestingly, the protein levels of most AMPs were similar in AD L and PsV L skin (Fig. 2). In contrast, LL-37 and CCL20 were highly and significantly upregulated in PsV L vs AD L skin, while ADM showed a similar trend with prominently higher levels in PsV L vs AD L (Fig. 2).
DISCUSSION
AMPs play a prominent role in the pathogenesis of several immune-mediated skin diseases. In PsV and rosacea, the production of these molecules is highly induced in keratinocytes, and some AMPs play an initial role in disease pathogenesis (5, 8, 9).
The impaired skin permeability barrier has been demonstrated as a major disease-driver factor in AD (10–12). The literature also suggests that the regulation of the skin permeability and antimicrobial barriers is closely connected (3, 13, 14). However, the examination of AMPs in AD is incomplete, and many discrepancies exist regarding the extent of AMP expression. These may partly be because AMP expression has often been compared with PsV rather than normal controls (Table II). In addition, in many cases, AMPs have been studied only at the mRNA level, which can be misleading, as proteins are the functional forms of the molecules. In most cases, studies did not include AD NL skin samples, which could provide critical data concerning the initial steps of disease pathophysiology (Table II). Lastly, to date, no study has investigated AMPs in AD by considering which functional subgroup each AMP belongs to.
| AD NL vs healthy | AD L vs healthy | PsV L vs AD L | ||||
| mRNA (RT-qPCR, MA, RNAseq) | protein (TS, IHC, WF) | mRNA (RT-qPCR, MA, RNAseq) | protein (TS, IHC, WF) | mRNA (RT-qPCR, MA, RNAseq) | protein (TS, IHC, WF) | |
| hBD-1 | nd | nd | ↓ in AD L (RT-qPCR) (24) no difference (RT-qPCR) (18) ↑ in AD L (RT-qPCR) (25) |
nd | NS (RT-qPCR) (18, 25) ↓ in AD L (MA) (18, 26) |
nd |
| hBD-2 | NS (RT-qPCR) (27) NS (TS/RT-qPCR) (28) ↑ AD NL (TS/RNAseq) (28) ↑ in AD NL (TS-RT-qPCR) (29) |
NS (IHC) (30) NS (TS/WB) (31) NS (TS/ELISA) (32, 33) NS (WF) (30) |
↑ in AD L (RT-qPCR) (18, 24, 25) NS (RT-qPCR) (34, 35) ↑ AD L (TS/RT-qPCR) (28, 29) ↑ AD L (TS/RNAseq) (28) |
↑ in AD L (IHC) (30, 35) NS (IHC) (18, 19, 34) ↑ in AD L (TS/ELISA) (31–33) ↑ AD L (TS/WB) (31) ↑ in AD L (WF) (30) |
↓ in AD L (RT-qPCR) (18, 24, 35, 36) ↓ in AD L (MA) (18, 26, 37) ↓ AD L (TS/RT-qPCR) (28) ↓ AD L (TS/RNAseq) (28) NS (RT-qPCR) (27) |
↓ in AD L (IHC) (18, 19, 30, 35) NS (TS) (31) ↓ in AD L (WF) (30) |
| hBD-3 | NS (RT-qPCR) (27) | ↑ in AD NL (IHC) (30) NS (TS/ELISA) (33) not detectable (WF) (30) |
↑ in AD L (RT-qPCR) (18, 25) NS (RT-qPCR) (24, 26, 36) |
↑ in AD L (IHC) (30) NS (IHC) (18, 26) NS (TS/ELISA) (33) not detectable (WF) (30) |
↓ in AD L (RT-qPCR) (26) NS (RT-qPCR) (18, 25, 27) NS (MA) (18) |
↓ in AD L (IHC) (26, 30) NS (IHC) (18) not detectable (WF) (30) |
| hBD-4 | nd | nd | nd | nd | nd | nd |
| LL-37 | NS (RT-qPCR) (27, 36, 38, 39) NS (TS/RT-qPCR) (28) ↑ AD NL (TS/RNAseq) (28) NS (TS/RT-qPCR) (29) |
NS (IHC) (38) not detectable (TS/ELISA) (33) |
↑ in AD L (RT-qPCR) (38) NS (RT-qPCR) (18, 24, 25, 34-36, 39) ↑ AD L (TS/RT-qPCR) (28, 29) ↑ AD L (TS/RNAseq) (28) |
↑in AD L (IHC) (38) NS (IHC) (19, 36) NS (WB) (34) not detectable (TS/ELISA) (33) NS (IF) (34) not detectable (WB) (19) |
↓ in AD L (RT-qPCR) (19, 35, 36, 39) NS (RT-qPCR) (18, 25, 27) NS (MA) (18) NS (TS/RT-qPCR) (28) NS (TS/RNAseq) (28) |
↓ in AD L (IHC) (19) |
| RNase7 | nd | NS (TS/ELISA) (33) NS (WF) (30) NS (IHC) (30) |
↑ in AD L (RT-qPCR) (25) | NS (IHC) (30) NS (TS/ELISA) (33) ↑ in AD L (WF) (30) |
↑ in AD L (RT-qPCR) (25) | NS (IHC) (30) NS (WF) (30) |
| Psoriasin | ↑ AD NL (TS/RNAseq) (28) | ↑ in AD NL (IHC) (40) NS (TS/ELISA) (33) ↑ in AD NL (WF) (30, 40) NS (IHC) (30) |
↑ in AD L (RT-qPCR) (25) NS (RT-qPCR) (35) ↑ AD L (TS/RNAseq) (28) |
↑ in AD L (IHC) (30, 35, 40) NS (TS/ELISA) (33) ↑ in AD L (WF) (30, 40) |
↓ in AD (RT-qPCR) (18) NS (RT-qPCR) (25) ↓ in AD L (MA) (18) NS (TS/RNAseq) (28) |
↓ in AD L (IHC) (30, 35, 40) ↓ in AD L (WF) (30) |
| Dermcidin | nd | nd | nd | nd | nd | nd |
| RNase5 | nd | nd | nd | nd | nd | nd |
| ADM | nd | nd | nd | nd | nd | nd |
| Elafin | ↑ AD NL (TS/RT-qPCR) (28) ↑ AD NL (TS/RNAseq) (28) |
nd | ↑ AD L (RT-qPCR) (18) NS (RT-qPCR) (35) ↑ AD L (TS/RT-qPCR) (28) ↑ AD L (TS/RNAseq) (28) |
NS (IHC) (18) | ↓ in AD L (RT-qPCR) (35) NS (RT-qPCR) (18) ↓ in AD L (MA) (18, 37) ↓ AD L (TS/RNAseq) (28) |
↓ in AD L (IHC) (18) ↓ AD L (TS/RT-qPCR) (28) |
| SLPI | nd | nd | nd | NS (IHC) (18) | nd | ↓ in AD L (IHC) (18) |
| LYZ | nd | nd | nd | nd | nd | nd |
| CCL20 | ↑ AD NL (TS/RT-qPCR) (28, 29) ↑ AD NL (TS/RNAseq) (28) |
nd | ↑ AD L (TS/RT-qPCR) (28, 29) ↑ AD L (TS/RNAseq) (28) |
NS (IHC) (35) | ↓ AD L (TS/RT-qPCR) (28) ↓ AD L (TS/RNAseq) (28) |
↓ in AD L (IHC) (35) |
| S100A8 | ↑ AD NL (TS/RNAseq) (28) | nd | ↑ AD L (RT-qPCR) (18) ↑ AD L (TS/RNAseq) (28) |
↑ AD L (IHC) (18) | ↓ in AD L (MA) (18) NS (RT-qPCR) (18) NS (TS/RNAseq) (28) |
NS (IHC) (18) |
| S100A9 | ↑ AD NL (TS/RT-qPCR) (28, 29) ↑ AD NL (TS/RNAseq) (28) |
nd | ↑ AD L (RT-qPCR) (18) ↑ AD L (TS/RT-qPCR) (28, 29) ↑ AD L (TS/RNAseq) (28) |
↑ AD L (IHC) (35) | ↓ in AD L (RT-qPCR) (18) ↓ in AD L (MA) (18) ↓ AD L (TS/RT-qPCR) (28) ↓ AD L (TS/RNAseq) (28) |
↓ in AD L (IHC) (35) |
| LCN2 | NS (TS/RNAseq) (28) | nd | NS (RT-qPCR) (35) NS (TS/RNAseq) (28) |
NS (IHC) (35) | ↓ in AD L (RT-qPCR) (35) ↓ AD L (TS/RNAseq) (28) ↓ AD L (MA) (37) |
↓ in AD L (IHC) (35) |
| Arrows indicate direction of change. | ||||||
| AD L: lesional atopic dermatitis skin; AD NL: non-lesional AD skin; IHC: immunohistochemistry; MA: microarray; nd: not determined; NS: no significant difference; PsV L: lesional psoriatic skin; TS: tape-stripping; WB: Western blot; WF: washing fluid; nd: not detected; hBD: human beta defensin; ADM, adrenomedullin; SLPI: secretory leukocyte Peptidase inhibitor; LYZ: lysozime; CCL20: C-C Motif Chemokine Ligand 20; S100A: S100 calcium-binding protein A; LCN2: lipocalin 2. | ||||||
The current study comprehensively analysed the 5 functional AMP groups, both at the mRNA and protein levels. AD NL (clinically asymptomatic) and AD L (clinically symptomatic) skin of patients with AD were compared with that of healthy controls. PsV is a chronic inflammatory skin disease with clinical and immunological characteristics which are completely distinct from those of AD; barrier damage is probably not the driver of this disease, thus this study also examined PsV L samples as a diseased control.
When comparing AMP levels in AD NL and control samples, no prominent difference was detected at the mRNA level, and only 2 AMPs showed significant alteration. At the protein level, only LL-37 showed alteration, as it was significantly decreased in AD NL. Limited data are available on AMPs’ protein expression in AD NL skin, and no studies have covered most AMPs at the same time. In line with the current findings, no prominent differences have been detected in AD NL vs controls (Table II).
Several AMPs were significantly altered at the mRNA level in AD L skin, and most AMPs were elevated compared with controls. At the protein level, AMP expression was prominently increased in AD L skin compared with controls, while 2 AMPs were unchanged and LL-37 was the only AMP with a highly decreased level in AD L skin. The published data are contradictory, despite the relatively high number of studies. Review articles emphasize that AMP expression is generally decreased in AD L skin; however, original studies showed predominantly elevated or unchanged AMP levels (Table II). Most data are available at the mRNA level, and the quantification of IHC results was either missing or subjective in most cases. Regarding acute AD, limited studies are available, which suggest that AMPs (S100A7, hBD-2, RNase7) are already highly induced and are not substantially enhanced in the acute-chronic transition (Table II).
When comparing AD L with PsV L samples, most AMPs were prominently induced in both diseases, with similar expression patterns; however, in many cases, AMP levels in AD did not reach the levels found in PsV. LL-37 showed the most striking difference between the 2 diseases, as the level was significantly increased in PsV lesions, but significantly decreased in AD skin vs controls. In line with the current results, the elevation of most AMPs in both diseases has also been demonstrated in other studies (Table II).
Reviewing the literature, data related to LL-37 in AD appear to be controversial. Only 1 study showed that LL-37 levels are higher in AD L compared with controls (Table II); however, the authors were unable to detect LL-37 by IHC in several AD L samples, which coincide with the current findings. Some investigations failed to detect significant differences in the LL-37 protein levels between AD L and controls (Table II). Finally, in several studies, LL-37 was below the limit of detection, in agreement with the current finding (Table II).
The finding in the current study that LL-37 was the only decreased AMP raises the question of whether LL-37 may play an important role in the pathogenesis of AD. Interestingly, literature data suggest that LL-37 is associated with all 3 major pathogenetic features of AD, including barrier damage, Staphylococcal hypercolonization, and Th2 inflammation. Under healthy conditions, a homeostatic balance is established between LL-37, the permeability barrier, and the microbiota, which maintains a non-inflammatory T cell (effector and resident memory) and Treg environment. However, literature data suggest that permeability barrier damage is strongly connected with LL-37 loss and Staphylococcal overgrowth. Decreased LL-37 levels lead to weakened tight junctions and impaired skin barrier function, since LL-37 is known to enhance the expression of tight junction molecules (e.g. claudins) that maintain skin permeability barrier (15). In addition, Staphylococcal overgrowth can damage both the antimicrobial and permeability barriers, as the cysteine protease (EcpA) activity of S. epidermidis was demonstrated to cleave LL-37 and a major desmosome component DSG1 in vitro (16). These findings are even more important considering that Staphylococcal density is substantially increased even on AD NL skin (17). Moreover, LL-37 loss may increase the susceptibility to Staphylococcus hypercolonization in AD (5, 18, 19), since LL-37 is highly effective and more potent against Staphylococcus species and biofilms than other AMPs, such as hBDs (20–22). In addition, during barrier damage, keratinocytes produce alarmins; mediators that initiate the promotion of Th2 cells. Finally, AD-specific Th2 cytokine milieu is known to inhibit the induction of LL-37 in vitro, which is in line with the decreased LL-37 levels in AD lesions in the current study in situ (11, 23).
In conclusion, AMPs were generally unchanged or increased in AD lesions compared with controls. The lack of LL-37 induction was the only impairment related to AMPs at the protein level in AD. The pattern and extent of AMP expression showed remarkable similarities in AD and PsV, except for LL-37. A prominent role of LL-37 in AD pathogenesis can be easily envisioned, as LL-37 is associated with all 3 major pathogenetic features of AD, including barrier damage, Staphylococcal hypercolonization, and Th2 inflammation (Fig. 3). The significantly decreased levels of LL-37 in AD NL skin indicate that LL-37 may play a driver role in the pathogenesis of AD, and raises the potential of LL-37 as a therapeutic target in the treatment of AD.
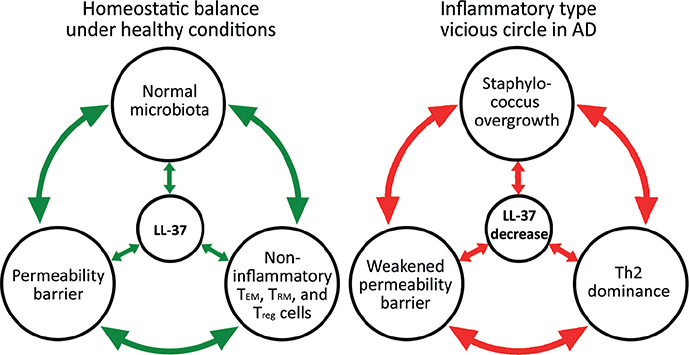
Fig. 3. The possible link between the 3 main pillars of atopic dermatitis (AD) pathogenesis. Left panel: Under steady-state conditions, the permeability barrier, the antimicrobial barrier, and the microbiota colonizing the skin (including Staphylococcus species) are in dynamic equilibrium. The homeostatic LL-37 level prevents the overgrowth of Staphylococcus species, and the intact permeability barrier limits their penetration into the skin. In addition, LL-37 enhances the function of the permeability barrier by promoting the expression of tight junction molecules. This homeostatic environment maintains a non-inflammatory effector memory (TEM), resident memory T cell (TRM), and Treg milieu. Right panel: Based on that LL-37 was the only AMP that showed significantly reduced levels already in non-lesional AD skin and in a disease-specific manner, we propose that LL-37 decrease could be one of the initial steps of disease pathophysiology. LL-37 loss leads to an increased ratio of Staphylococcus species. Staphylococcal proteolytic activity cleaves LL-37 and the desmosome component DSG1 protein, which can further enhance Staphylococcal colonization. As LL-37 levels decrease, positive feedback from the permeability barrier-maintaining capacity of LL-37 is abolished, resulting in a decreased expression of molecules composing tight junctions. This leads to further barrier damage, which promotes the hypercolonization of Staphylococcus species. Barrier damage also promotes alarmin production by keratinocytes, which mediators initiate the maturation and proliferation of inflammatory Th2 cells, leading to a vicious circle.
ACKNOWLEDGEMENTS
The publication was supported by Hungarian Research Grants (NKFIH K-128250, NKFIH K-142348, and NKFIH PD-131689) and the EFOP-3.6.1-16-2016-00022 project. The project was co-financed by the European Union and the European Social Fund. This project was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences (ZsD) and the ÚNKP-22-3 New National Excellence Program of the Ministry for Culture and Innovation from the source of the national research, development and innovation fund (LSz).
REFERENCES
- Nguyen HLT, Trujillo-Paez JV, Umehara Y, Yue H, Peng G, Kiatsurayanon C, et al. Role of antimicrobial peptides in skin barrier repair in individuals with atopic dermatitis. Int J Mol Sci 2020; 21.
- Beke G, Dajnoki Z, Kapitany A, Gaspar K, Medgyesi B, Poliska S, et al. Immunotopographical differences of human skin. Front Immunol 2018; 9: 424.
- Borkowski AW, Gallo RL. The coordinated response of the physical and antimicrobial peptide barriers of the skin. J Invest Dermatol 2011; 131: 285–287.
- Braff MH, Gallo RL. Antimicrobial peptides: an essential component of the skin defensive barrier. Curr Top Microbiol Immunol 2006; 306: 91–110.
- Niyonsaba F, Kiatsurayanon C, Chieosilapatham P, Ogawa H. Friends or foes? Host defense (antimicrobial) peptides and proteins in human skin diseases. Exp Dermatol 2017; 26: 989–998.
- Jenei A, Dajnoki Z, Medgyesi B, Gaspar K, Beke G, Kinyo A, et al. Apocrine gland-rich skin has a non-inflammatory IL-17-related immune milieu, that turns to inflammatory IL-17-mediated disease in hidradenitis suppurativa. J Invest Dermatol 2019; 139: 964–968.
- Jenei A, Kallo G, Dajnoki Z, Gaspar K, Szegedi A, Kapitany A, et al. Detection of antimicrobial peptides in stratum corneum by mass spectrometry. Int J Mol Sci 2021; 22.
- Yamasaki K, Gallo RL. Antimicrobial peptides in human skin disease. Eur J Dermatol 2008; 18: 11–21.
- Clausen ML, Slotved HC, Krogfelt KA, Andersen PS, Agner T. In vivo expression of antimicrobial peptides in atopic dermatitis. Exp Dermatol 2016; 25: 3–9.
- Yang G, Seok JK, Kang HC, Cho YY, Lee HS, Lee JY. Skin barrier abnormalities and immune dysfunction in atopic dermatitis. Int J Mol Sci 2020; 21.
- Beck LA, Cork MJ, Amagai M, De Benedetto A, Kabashima K, Hamilton JD, et al. Type 2 inflammation contributes to skin barrier dysfunction in atopic dermatitis. JID Innov 2022; 2: 100131.
- Egawa G, Kabashima K. Multifactorial skin barrier deficiency and atopic dermatitis: Essential topics to prevent the atopic march. J Allergy Clin Immunol 2016; 138: 350–358 e351.
- Dorschner RA, Pestonjamasp VK, Tamakuwala S, Ohtake T, Rudisill J, Nizet V, et al. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J Invest Dermatol 2001; 117: 91–97.
- Aberg KM, Man MQ, Gallo RL, Ganz T, Crumrine D, Brown BE, et al. Co-regulation and interdependence of the mammalian epidermal permeability and antimicrobial barriers. J Invest Dermatol 2008; 128: 917–925.
- Akiyama T, Niyonsaba F, Kiatsurayanon C, Nguyen TT, Ushio H, Fujimura T, et al. The human cathelicidin LL-37 host defense peptide upregulates tight junction-related proteins and increases human epidermal keratinocyte barrier function. J Innate Immun 2014; 6: 739–753.
- Cau L, Williams MR, Butcher AM, Nakatsuji T, Kavanaugh JS, Cheng JY, et al. Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J Allergy Clin Immunol 2021; 147: 955–966 e916.
- Tauber M, Balica S, Hsu CY, Jean-Decoster C, Lauze C, Redoules D, et al. Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J Allergy Clin Immunol 2016; 137: 1272–1274 e1273.
- de Jongh GJ, Zeeuwen PL, Kucharekova M, Pfundt R, van der Valk PG, Blokx W, et al. High expression levels of keratinocyte antimicrobial proteins in psoriasis compared with atopic dermatitis. J Invest Dermatol 2005; 125: 1163–1173.
- Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med 2002; 347: 1151–1160.
- Noore J, Noore A, Li B. Cationic antimicrobial peptide LL-37 is effective against both extra- and intracellular Staphylococcus aureus. Antimicrob Agents Chemother 2013; 57: 1283–1290.
- Kang J, Dietz MJ, Li B. Antimicrobial peptide LL-37 is bactericidal against Staphylococcus aureus biofilms. PLoS One 2019; 14: e0216676.
- Midorikawa K, Ouhara K, Komatsuzawa H, Kawai T, Yamada S, Fujiwara T, et al. Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infect Immun 2003; 71: 3730–3739.
- Howell MD, Gallo RL, Boguniewicz M, Jones JF, Wong C, Streib JE, et al. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity 2006; 24: 341–348.
- Gambichler T, Skrygan M, Tomi NS, Altmeyer P, Kreuter A. Changes of antimicrobial peptide mRNA expression in atopic eczema following phototherapy. Br J Dermatol 2006; 155: 1275–1278.
- Gambichler T, Skrygan M, Tomi NS, Othlinghaus N, Brockmeyer NH, Altmeyer P, et al. Differential mRNA expression of antimicrobial peptides and proteins in atopic dermatitis as compared to psoriasis vulgaris and healthy skin. Int Arch Allergy Immunol 2008; 147: 17–24.
- Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol 2003; 112: 1195–1202.
- Hata TR, Kotol P, Boguniewicz M, Taylor P, Paik A, Jackson M, et al. History of eczema herpeticum is associated with the inability to induce human beta-defensin (HBD)-2, HBD-3 and cathelicidin in the skin of patients with atopic dermatitis. Br J Dermatol 2010; 163: 659–661.
- He H, Bissonnette R, Wu J, Diaz A, Saint-Cyr Proulx E, Maari C, et al. Tape strips detect distinct immune and barrier profiles in atopic dermatitis and psoriasis. J Allergy Clin Immunol 2021; 147: 199–212.
- Guttman-Yassky E, Diaz A, Pavel AB, Fernandes M, Lefferdink R, Erickson T, et al. Use of tape strips to detect immune and barrier abnormalities in the skin of children with early-onset atopic dermatitis. JAMA Dermatol 2019; 155: 1358–1370.
- Harder J, Dressel S, Wittersheim M, Cordes J, Meyer-Hoffert U, Mrowietz U, et al. Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. J Invest Dermatol 2010; 130: 1355–1364.
- Asano S, Ichikawa Y, Kumagai T, Kawashima M, Imokawa G. Microanalysis of an antimicrobial peptide, beta-defensin-2, in the stratum corneum from patients with atopic dermatitis. Br J Dermatol 2008; 159: 97–104.
- Clausen ML, Jungersted JM, Andersen PS, Slotved HC, Krogfelt KA, Agner T. Human beta-defensin-2 as a marker for disease severity and skin barrier properties in atopic dermatitis. Br J Dermatol 2013; 169: 587–593.
- Clausen ML, Slotved HC, Krogfelt KA, Agner T. Measurements of AMPs in stratum corneum of atopic dermatitis and healthy skin-tape stripping technique. Sci Rep 2018; 8: 1666.
- Goo J, Ji JH, Jeon H, Kim MJ, Jeon SY, Cho MY, et al. Expression of antimicrobial peptides such as LL-37 and hBD-2 in nonlesional skin of atopic individuals. Pediatr Dermatol 2010; 27: 341–348.
- Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, Zaba LC, Cardinale I, Nograles KE, et al. Low expression of the IL-23/Th17 pathway in atopic dermatitis compared to psoriasis. J Immunol 2008; 181: 7420–7427.
- Howell MD, Novak N, Bieber T, Pastore S, Girolomoni G, Boguniewicz M, et al. Interleukin-10 downregulates anti-microbial peptide expression in atopic dermatitis. J Invest Dermatol 2005; 125: 738–745.
- Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol 2003; 171: 3262–3269.
- Ballardini N, Johansson C, Lilja G, Lindh M, Linde Y, Scheynius A, et al. Enhanced expression of the antimicrobial peptide LL-37 in lesional skin of adults with atopic eczema. Br J Dermatol 2009; 161: 40–47.
- Mallbris L, Carlen L, Wei T, Heilborn J, Nilsson MF, Granath F, et al. Injury downregulates the expression of the human cathelicidin protein hCAP18/LL-37 in atopic dermatitis. Exp Dermatol 2010; 19: 442–449.
- Glaser R, Meyer-Hoffert U, Harder J. [Antimicrobial peptides in atopic dermatitis. A paradigm shift?]. Hautarzt 2009; 60: 761–762.