ORIGINAL ARTICLE
Genotyping of the rs1800440 Polymorphism in CYP1B1 Gene and the rs9258883 Polymorphism in HLA-B Gene in a Spanish Cohort of 223 Patients with Frontal Fibrosing Alopecia
David SACEDA-CORRALO1–3, Daniel ORTEGA-QUIJANO1,3, Gloria MUÑOZ-MARTÍN4, Óscar M. MORENO-ARRONES1,3, Cristina PINDADO-ORTEGA1,3, Tuntas RAYINDA5,6, Ana MELIÁN-OLIVERA1, Carlos AZCÁRRAGA-LLOBET1, Patricia BURGOS-BLASCO1, María ELENA CASTAÑEDA-BERMÚDEZ7, Francisco J. DEL CASTILLO4 and Sergio VAÑÓ-GALVÁN1,3,8
1Dermatology Department, University Hospital Ramón y Cajal, Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), Madrid, 2Departamento de Biología de Sistemas, Facultad de Medicina, Universidad de Alcalá, IRYCIS, 3Trichology Unit, Grupo de Dermatologia Pedro Jaén, 4UCA Translational Genomics, University Hospital Ramón y Cajal, Madrid, Spain, 5St John’s Institute of Dermatology, King’s College London, London, UK, 6Guy’s and St Thomas’ NHS Foundation Trust, London, UK, 7Jalisco Dermatological Institute, Guadalajara, Mexico and 8Departamento de Medicina, Facultad de Medicina, Universidad de Alcalá, IRYCIS, Madrid, Spain
The pathogenesis of frontal fibrosing alopecia has been linked to specific genetic variants. CYP1B1 codes for a component of the cytochrome p450 machinery that is involved in the metabolism of xenobiotic oestrogens. The study of the prevalence of polymorphisms in this gene may help to understand their role in the development of frontal fibrosing alopecia. The aim of this study is to describe the frequency of genetic variations in the alleles HLA-B*07:02 and CYP1B1 in patients with frontal fibrosing alopecia. A cross-sectional study was designed to evaluate blood samples from patients with frontal fibrosing alopecia who attended the Dermatology Department at University Hospital Ramón y Cajal (Madrid, Spain), in search of the polymorphisms rs9258883 and rs1800440 in the alleles HLA-B*07:02 and CYP1B1, respectively. A total of 223 patients were included in the study. Among the 83.8% of patients who carried the rs9258883 polymorphism in HLA-B*07:02, 58.7% were heterozygous for this variant and it was not present in 14.8% of the cases. The majority of patients with frontal fibrosing alopecia lacked the protective rs1800440 polymorphism in CYP1B1 (75.2%). This suggests a relevant role of this variant in development of frontal fibrosing alopecia. The genetic approach to this condition might influence patient prognosis and therapy options.
Key words: cicatricial alopecia; genetic loci; hair loss; HLA antigens; pathogenesis; xenobiotics.
SIGNIFICANCE
The origin of frontal fibrosing alopecia has been linked to 2 genetic variants in CYP1B1 and HLA-B*07:02. CYP1B1 plays a role in the metabolism of xenobiotic oestrogens and HLA-B is related to the aberrant immune response. This study includes 223 patients with frontal fibrosing alopecia and determines how frequent are the genetic variants described. A total of 83.8% of patients with frontal fibrosing alopecia carried the rs9258883 polymorphism in HLA-B*07:02, and the majority of them lacked the protective rs1800440 polymorphism in CYP1B1 (75.2%). The high proportion of both genetic variants in the group of patients in this study suggests a relevant role of genetics in the development of frontal fibrosing alopecia.
Citation: Acta Derm Venereol 2023; 103: adv9604. DOI: https://doi.org/10.2340/actadv.v103.9604.
Copyright: © Published by Medical Journals Sweden, on behalf of the Society for Publication of Acta Dermato-Venereologica. This is an Open Access article distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (https://creativecommons.org/licenses/by-nc/4.0/)
Accepted: Aug 15, 2023; Published: 18 Sep, 2023
Corr: Daniel Ortega Quijano, Dermatology Department, Ramon y Cajal Hospital. Carretera Colmenar Viejo km 9.100, ES-28034 Madrid, Spain. E-mail: danielortegaquijano@gmail.com
Competing interests and funding: The authors have no conflicts of interest to declare.
INTRODUCTION
Frontal fibrosing alopecia (FFA) is a scarring and inflammatory permanent hair loss condition involving frontotemporal hairline and eyebrows (1–3). FFA primarily affects women, and it has also been linked to various autoimmune illnesses, such as chronic discoid lupus erythematosus, autoimmune thyroid disease, and pernicious anaemia (4, 5). The aetiopathogenesis of FFA, however, is unknown (6, 7). Hundreds of cases of FFA have been documented in the literature since 1994, with a constant increase in its incidence worldwide. Moreover, genetic research has played a key role in identifying molecular pathways underpinning other hair loss disorders, such as alopecia areata (8, 9). FFA has been found among siblings and family members, implying a genetic link and familial disease was studied in detail by Tziotzios et al. (10). Thus, no highly penetrant alleles were identified to segregate in a Mendelian pattern, suggesting that the trait was polygenic or genetically complex (11).
A subsequent genome-wide association study (GWAS) examined common genetic variations in patients with FFA (12). In fact, this study evaluated more than 15 million common variants in 2 large cohorts of female patients with FFA and controls from the UK and Spain. Four causal variants were identified (HLA-B*07:02, CYP1B1, SEMA4B and ST3GAL1), with the largest effect size observed for HLA-B*07:02 and CYP1B1. The latter encodes a part of cytochrome P450, which is involved in oestrogen and xenobiotic metabolism, and it may well interact with hormonal exogenous exposures. The presence of these polymorphisms in diverse FFA populations might help us to better understand their impact on the disease’s aetiopathogenesis and clinical characteristics.The aim of this study is to determine the prevalence of the above variants in a large cohort of patients with FFA at University Hospital Ramón y Cajal and to correlate its genotype with clinical characteristics.
MATERIALS AND METHODS
Patients
The study included men and women over the age of 18 years with a confirmed diagnosis of FFA who attended the Dermatology Department at University Hospital Ramón y Cajalbetween January 2015 and September 2020. FFA was defined as trichoscopy-detected regression of the frontotemporal hairline and loss of follicular openings, as well as histological analysis of a skin biopsy. Samples and data from patients included in this study were provided by the Biobank Hospital Ramón y Cajal-IRYCIS (National Registry of Biobanks B.0000678), integrated in the Platform ISCIII Biobanks and Biomodels (PT20/00045) and they were processed following standard operating procedures with the appropriate approval of the Ethics and Scientific Committees at University Hospital Ramón y Cajal. All participants gave written informed consent allowing a blood sample to be stored. If the blood samples obtained were of low quality or had degraded, the patients were excluded from the study.
Study design
A cross-sectional observational study was conducted to evaluate blood samples for genetic variants at 2 genomic loci: 6p21.1 (gene HLA-B) and 2p22.2 (gene CYP1B1). The local institutional review board gave their approval to the research (Code: GEN-AFF 04-2019).
Data on epidemiology, comorbidities, personal gynaecological background and clinical presentation were collected. After at least 1 year of treatment, a satisfactory response to treatment was defined as no hair loss progression in the frontal and lateral hairline, according to the Frontal Fibrosing Alopecia Severity Score (FFASS) (13).
Storage and genotyping
The patients’ blood samples were collected and processed into 250-µl cell rest aliquots, then stored at –80 °C of the Ramón y Cajal Hospital’s Biobank. DNA extraction and genotyping were performed at the Hospital’s Central Translational Genomics Support Unit (UCA-GT). The DNA samples extraction was obtained in the first stage using the Qiagen Flexigem DNA Extraction Kit (Qiagen, Hilden, Germany). Next, DNA was quantified with the nanodrop, the Qubit, and the Tape Station 2200 (Thermo Fisher Scientific, Waltham, MA, USA) to check its concentration and quality. The appropriate PCRs were run to amplify the 2 regions of interest containing the 2 polymorphisms. Electrophoresis has been used to confirm PCR amplification. The PCRs were purified using the ExoSAP-IT kit (Thermo Fisher Scientific, Waltham, MA, USA), and Sanger sequencing (Macrogen Inc., Seoul, Republic of Korea) of both fragments was conducted to genotype the 2 single-nucleotide polymorphisms (SNPs) in each patient. For the rs1800440 polymorphism located in the CYP1B1 gene, the genotypes to be considered would be AA, AG or GG. While for the rs9258883 polymorphism located in the HLA-B locus, the possible genotypes would be CC, CT or TT.
Statistical analysis
All analyses were carried out using the statistical software package SPSS 25.0. (IBM SPSS Statistics for Macintosh; IBM Corp., Armonk, NY, USA). For the patients’ demographic and clinical features, descriptive statistics were employed (mean and range for continuous measures, and frequency and percentage for categorical variables).
A bivariate analysis was used to compare groups, employing the χ2 test or, where applicable, the Fisher’s exact test. According to the normal distribution of the data, the Student’s t-test or the Kruskal–Wallis test was utilized to compare quantitative variables.
RESULTS
Patients
A total of 223 patients who agreed to participate were enrolled into the study (Fig. 1). There were 221 women and 2 men, with a mean age of 61 (range 32–86) years. All patients were Caucasian. Family history was confirmed in 20 patients (9.0%), while in 6 of them there was a first-grade relative with FFA. Thyroid alterations were present in 80.0% of patients, with hypothyroidism being the most frequent condition (Hashimoto’s disease in 10.7%). Table I summarizes the medical comorbidities, gynaecological background, and hormonal therapy received by patients.
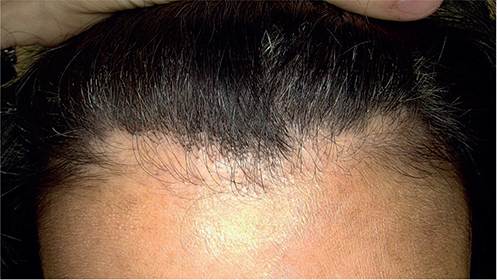
Fig. 1. Clinical presentation of hair loss in a patient with frontal fibrosing alopecia with a pseudo-fringe pattern.
Clinical patterns in frontal fibrosing alopecia
The clinical characteristics of FFA were analysed in all patients. The 3 patterns of hair loss in FFA were detected among the patients included (Fig. 2). Pattern I “linear” was observed in 87/203 patients (42.9%), pattern II “zig-zag” in 65/203 patients (32.0%) and pattern III “pseudo-fringe” in 44/203 patients (21.7%). Seven patients (7/203, 3.4%) had a clinical pattern that was not clearly defined. Loss of eyebrows was total in 78/196 patients (39.8%) and partial in 44/196 patients (22.4%). In contrast, no eyebrows loss was observed in 74/196 patients (37.8%). Facial papules were present in 56/196 cases (28.6%). Inflammatory signs were detected in 92/203 patients (45.3%).
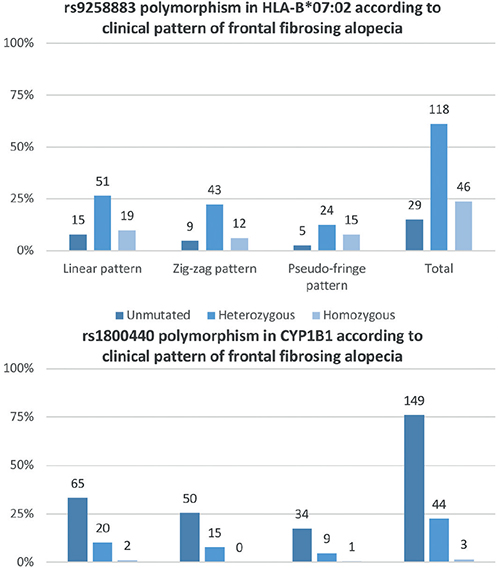
Fig. 2. Distribution of genetic variations on the alleles identified in the genome-wide association study (GWAS) in the current study sample. The prevalence of rs1800440 polymorphism in CYP1B1 is remarkably low and distribution of the rs9258883 variant of HLA-B*07:02 allele is more heterogeneous.
The response to medical treatment was evaluated in 107 patients (47.9%) after a 1-year follow-up. The alopecia was stabilized in 60 of them (56.1%), on the frontal and lateral hairline. All patients underwent the same therapy, including oral dutasteride (3–7 capsules per week), topical clobetasol propionate 0.05% twice weekly, and topical minoxidil 5% 5 nights per week.
Genotyping
Genetic analysis of blood samples was carried out for HLA-B*07:02 and for CYP1B1. The HLA-B*07:02 allele presented the rs9258883 polymorphism in 187 individuals (83.8%), 56 (25.1%) of whom were homozygous, while 131 (58.7%) were heterozygous. In comparison, 33 individuals (14.8%) lacked the described variant in this locus, and in 3 patients (1.3%) the allele could not be assessed due to the low quality of DNA extracted. Singularly, most FFA patients did not present the protective mutation in CYP1B1 (167 patients, 75.2%). However, 55 patients (24.8%) showed the rs180040 polymorphism. Fifty-one of them were heterozygous and just 4 patients were homozygous (23.0% and 1.8%, respectively). In only 1 patient the variant could not be studied, also due to the low quality of DNA extracted. The relationship between mutations in both loci is shown in Table II.
On the whole, no statistically significant differences were found between the clinical pattern of hair loss and genetic mutations (Fig. 3). Moreover, no other statistically significant associations were observed between the genetic mutations identified and the patients’ clinical characteristics or treatment responses.
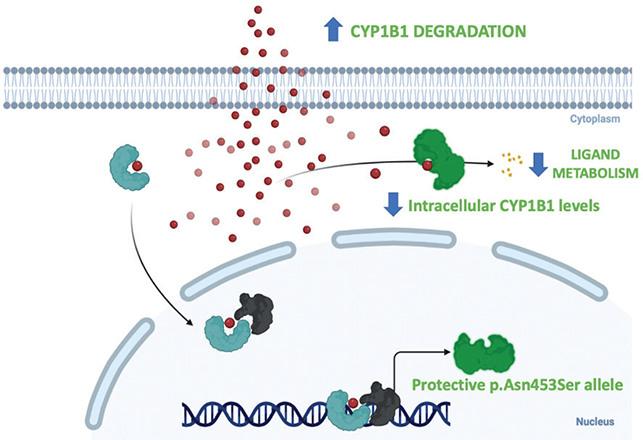
Fig. 3. The protective p.Asn453Ser allele enhances the rate of CYP1B1 degradation, resulting in lower intracellular CYP1B1 levels. Individuals with this allele are protected from developing frontal fibrosing alopecia.
DISCUSSION
A genetic explanation for FFA has been hypothesized for a decade, and there have been some early attempts to investigate it, particularly in short publications and case reports (14). As an illustration, Navarro-Belmonte et al. (15) identified 4 FFA-affected families and 8 mother–daughter cases, implying that the gene anticipation phenomenon was associated to the onset of FFA in daughters at an earlier age. Furthermore, researchers identified a 67% concordance in 2 pairs of FFA monozygotic twins, which was greater than in comparable disorders, such as systemic lupus erythematosus (16). In these trials, however, the small sample size was deemed insufficient to show a link between the genome and the FFA. But it was not until Tziotzios et al.’s (12) latest study that we observed an in-depth genetic picture in patients with FFA.
Human major histocompatibility complex research in patients with FFA is still in its early stages. In essence, HLA A, HLA B, and HLA C expression is low in the hair follicle bulge area and the outer root sheath. As mentioned at the outset, the identified HLA-B*07:02 mutation in the GWAS study (HLA class I), has been postulated to assist in the presentation of autoantigens, facilitating the auto-inflammatory lymphocytic death of the epithelial hair follicle stem cells located in the hair follicle bulge. Past reports have suggested that the collapse of hair follicles, the disease’s hallmark sign, might be caused by changes in the expression pattern of HLA class I and class II molecules (17). One argument favouring this is the study by Ramos et al., in which 2 susceptible HLA class I haplotypes were identified (HLA-C*17:01:01:02/B*42:01:01:01 and C*07:02:01:03/B*07:02:01:01) in a Brazilian large familiar cluster with FFA (14). Similarly, research in 13 cases of FFA in Spain uncovered a linkage between the F16A HLA class I haplotype and the CYP21A2 gene p.V281L mutation (4). Chan et al. (18) on the other hand, found no HLA-DR1 association (HLA class II) in 2 siblings with FFA. Nonetheless, these studies have several limitations that make it difficult to draw significant conclusions. As the HLA area is dense and complex, only in-depth genetic research can provide insight into its relevance in this disorder.
The association found in the GWAS study is much more solid than those described in smaller studies. Authors suggested that the variant in the allele HLA-B*07:02 can induce a 5-fold increase in the risk of FFA. This variant was found in most of the patients in this study, although the most common presentation was a heterozygous mutation in the patients’ allele (58.7%). However, this gene mutation was not observed in 14.8% of subjects, raising the question that distinct HLA alleles may be involved in the pathogenesis of the disease.
As previously noted, CYP1B1 gene variations accounted for the large number of FFA cases in the current study sample. This gene has attracted attention because of its unusual properties and characteristics (19). The CYP1B1 gene belongs to the CPYP1 gene family, including the CYP1A1 and CYP1A2 genes (20). It is expressed in the liver and extrahepatic tissue and is located on chromosome 2 (21). It does, however, perform xenobiotic metabolism, which includes the metabolic activation of polycyclic aromatic hydrocarbons via the microsomal enzyme cytochrome B450 1B1 (22). This enzyme aids in the conversion of oestradiol and oestrone to their hydroxylated catechol oestrogens through oxidative metabolism (23). Previous research, observed a possible role for intracellular CY1PB1 levels in the risk of carcinogenity (19, 20). CYP1B1 is regulated by several key transcription factors, such as the aryl hydrocarbon receptor (AhR) (24). The activation of this signalling pathway suggests that it plays a crucial role in the development of immunological, toxicological, and oncological processes. It is also an important regulator of skin barrier function (25). Interestingly, the interaction between dioxin-like substances and the AhR may be related to the suppression of PPAR-gamma, an aetiopathogenic mechanism that has been described in lichen planopilaris (26). The possible role of AhR on occurrence of FFA is still to be elucidated, although a higher expression of this receptor has been detected in skin biopsies from patients with FFA (27).
Only 4 patients presented a homozygous mutation in the CYP1B1 gene that was related to a protective allele. In fact, most of the study patients (75.2%) had not the rs180040 polymorphism described, as expected. The presence of the protective allele expressed in homozygous might be an important predictor factor for low-risk of FFA. However, larger studies must corroborate the extremely low prevalence of this genetic variation in FFA population. The FFA protective p.Asn453Ser allele enhances the rate of CYP1B1 degradation, resulting in lower intracellular CYP1B1 levels, according to functional analysis of allelic variation in CYP1B1 (28). Considering the importance of CYP1B1 in sex hormone metabolism, FFA’s female preponderance, and its worldwide rise in occurrence, FFA is most likely the consequence of increased female exposure to a CYP1B1 substrate (Fig. 3) (12).
The current study has several limitations. First, we selectively genotype 2 particular SNPs in the sample, excluding additional loci that may well be involved in FFA. The study’s major goal, however, was to characterize the genetic variations associated with this type of alopecia that had previously been observed. Secondly, we included only a limited number of particular cases of FFA, comprising 2 males who had been diagnosed with the condition. The actual data could become more heterogeneous as a result of this. Future research should focus on the genetic bases of FFA in these patients. We also added 6 relatives, which increased the likelihood of detecting similar mutations in most of them. Thirdly, 139 individuals out of the total of the patients included in the study had already been investigated in the previous GWAS. Despite this, describing the genetic variations in a larger sample of patients with FFA might aid in determining the prevalence of these molecular findings in an ongoing cohort in clinical practice. Lastly, the retrospective design of the study entails some missing clinical data. However, the missing data is few and does not significantly impact the data analysis.
In a nutshell, this is the first description of a large Spanish FFA sample based on the genetic findings in the GWAS study. The above-mentioned aspects suggest that HLA class I mutations, and the CYP1B1 gene in particular, could play a decisive role in the development of FFA. The genetic approach to this disease may have an impact on prognosis and on future therapies for patients.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the patients and the BiobankHospital Ramón y Cajal-IRYCIS (B.0000678), integrated in the Platform ISCIII Biobanks and Biomodels (PT20/00045) for their collaboration.
This research was funded by a grant from Biomedical Research Institute IRyCIS.
The current study has been approved by an Institutional Review Board at University Hospital Ramón y Cajal (study number: GEN AFF 04-2019). The patients in this study have given written informed consent to publication of their case details.
REFERENCES
- Kossard S, Lee MS, Wilkinson B. Postmenopausal frontal fibrosing alopecia: a frontal variant of lichen planopilaris. J Am Acad Dermatol 1997; 36: 59–66.
- Kossard S. Postmenopausal frontal fibrosing alopecia. Scarring alopecia in a pattern distribution. Arch Dermatol 1994; 130: 770–774.
- Iorizzo M, Tosti A. Frontal fibrosing alopecia: an update on pathogenesis, diagnosis, and treatment. Am J Clin Dermatol 2019; 20: 379–390.
- Porriño-Bustamante ML, López-Nevot MÁ, Aneiros-Fernández J, Casado-Ruiz J, García-Linares S, Pedrinacci-Rodríguez S, et al. Study of human leukocyte antigen (HLA) in 13 cases of familial frontal fibrosing alopecia: CYP21A2 gene p.V281L mutation from congenital adrenal hyperplasia linked to HLA class I haplotype HLA-A*33:01; B*14:02; C*08:02 as a genetic marker. Australas J Dermatol 2019; 60: e195–e200.
- McSweeney SM, Christou EAA, Dand N, Boalch A, Holmes S, Harries M. et al. Frontal fibrosing alopecia: a descriptive cross-sectional study of 711 cases in female patients from the UK. Br J Dermatol 2020; 183: 1136–1138.
- Dlova N, Goh C-L, Tosti A. Familial frontal fibrosing alopecia. Br J Dermatol 2013; 168: 220–222.
- Photiou L, Nixon RL, Tam M, Green J, Yip L. An update of the pathogenesis of frontal fibrosing alopecia: what does the current evidence tell us? Australas J Dermatol 2019; 60: 99–104.
- Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 2010; 466: 113–117.
- Kanti V, Constantinou A, Reygagne P, Vogt A, Kottner J, Blume-Peytavi U. Frontal fibrosing alopecia: demographic and clinical characteristics of 490 cases. J Eur Acad Dermatology Venereol 2019; 33: 1976–1983.
- Tziotzios C. Molecular exploration of frontal fibrosing alopecia – doctoral thesis. 2019. [accessed 2022 Jun 14]. Available from https://kclpure.kcl.ac.uk/portal/en/theses/molecular-exploration-of-frontal-fibrosing-alopecia(5a7dbdd0-ac16-44a7-916d-fee3fd0eebfa).html.
- Tziotzios C, Stefanato CM, Fenton DA, Simpson MA, McGrath JA. Frontal fibrosing alopecia: reflections and hypotheses on etiology and pathogenesis. Exp Dermatol 2016; 25: 847–852.
- Tziotzios C, Petridis C, Dand N, Ainali C, Saklatvala JR, Pullabhatla V, et al. Genome-wide association study in frontal fibrosing alopecia identifies four susceptibility loci including HLA-B*07:02. Nat Commun 2019; 10: 1–9.
- Saceda-Corralo D, Moreno-Arrones ÓM, Fonda-Pascual P, et al. Development and validation of the Frontal Fibrosing Alopecia Severity Score. J Am Acad Dermatol 2018; 78: 522–529.
- Ramos PM, Garbers LEFM, Silva NSB, Castro CFB, Andrade HS, Souza AS, et al. A large familial cluster and sporadic cases of frontal fibrosing alopecia in Brazil reinforce known human leucocyte antigen (HLA) associations and indicate new HLA susceptibility haplotypes. J Eur Acad Dermatol Venereol 2020; 34: 2409–2413.
- Navarro-Belmonte MR, Navarro-López V, Ramírez-Boscà A, et al. Case series of familial frontal fibrosing alopecia and a review of the literature. J Cosmet Dermatol 2015; 14: 64–69.
- Tziotzios C, Fenton DA, Stefanato CM, McGrath JA. Familial frontal fibrosing alopecia. J Am Acad Dermatology 2015; 73: e37.
- Harries MJ, Meyer K, Chaudhry I, et al. Lichen planopilaris is characterized by immune privilege collapse of the hair follicle’s epithelial stem cell niche. J Pathol 2013; 231: 236–247.
- Chan D, Kartono F, Ziegler R, Abdulwahab N, DiPaola N, Flynn J WH, et al. Absence of HLA-DR1 positivity in 2 familial cases of frontal fibrosing alopecia. J Am Acad Dermatol 2014; 71: e208–e210.
- Faiq M, Dada R, Sharma R, Saluja D, Dada T. CYP1B1: a unique gene with unique characteristics. Curr Drug Metab 2014; 15: 893–914.
- Carrera AN, Grant MKO, Zordoky BN. CYP1B1 as a therapeutic target in cardio-oncology. Clin Sci 2020; 134: 2897–2927.
- Tang YM, Wo YYP, Stewart J, Hawkins AL, Griffin CA, Sutter TR, et al. Isolation and characterization of the human cytochrome p450 CYP1B1 gene. J Biol Chem 1996; 271: 28324–28330.
- Shimada T, Gillam EMJ, Sutter TR, Strickland PT, Guengerich FP, Yamazaki H. Oxidation of xenobiotics by recombinant human cytochrome P450 1B1. Drug Metab Dispos 1997; 25: 617–622.
- Sowers MFR, Wilson AL, Kardia SR, Chu J, McConnell DS. CYP1A1 and CYP1B1 polymorphisms and their association with estradiol and estrogen metabolites in women who are premenopausal and perimenopausal. Am J Med 2006; 119: S44–51.
- Zhou L. AHR function in lymphocytes: emerging concepts. Trends Immunol 2016; 37: 17–31.
- Fernández-Gallego N, Sánchez-Madrid F, Cibrian D. Role of AHR ligands in skin homeostasis and cutaneous inflammation. Cells 2021; 10: 1–27.
- Karnik P, Tekeste Z, McCormick TS, Gilliam AC, Price VH, Cooper KD, et al. Hair follicle stem cell-specific pparγ deletion causes scarring alopecia. J Invest Dermatol 2009; 129: 1243–1257.
- Doche I, Pagliari C, Hordinsky MK, Wilcox GL, Rivitti-Machado MCM, Romiti R, et al. Overexpression of the aryl hydrocarbon receptor in frontal fibrosing alopecia and lichen planopilaris: a potential pathogenic role for dioxins?: an investigational study of 38 patients. J Eur Acad Dermatology Venereol 2020; 34: e326–e329.
- Bandiera S, Weidlich S, Harth V, Broede P, Ko Y, Friedberg T. Proteasomal degradation of human CYP1B1: effect of the Asn453Ser polymorphism on the post-translational regulation of CYP1B1 expression. Mol Pharmacol 2005; 67: 435–443.